A comparison of approaches to accelerate NGS sample preparation and their amenability to automation
Traditional next-generation sequencing (NGS) sample preparation methods can be labor-intensive, requiring careful sample handling by experienced laboratory personnel to produce efficient and reliable sequencing results. Recent years have seen the development and adaptation of several approaches to sample preparation, aiming to improve speed, throughput, and reproducibility.
Many of these approaches are also compatible with automation to greater or lesser extents, providing the opportunity to minimize human error and variation, and further improve consistency between samples from different batches.
The need for sample throughput in NGS workflows
Sample throughput is a key factor in sample preparation for high-throughput sequencing labs, having a direct impact on productivity and cost-effectiveness.
What is sample throughput?
Sample throughput refers to the number of samples processed simultaneously (parallel processing) or in a given timeframe (per day, week, or longer).
NGS applications have varying throughput requirements. Small-scale/low-throughput sample preparation, such as that in small academic research projects, is usually manageable by manual processing, while higher throughput applications can require a level of automation, dependent on format, scale, and equipment options available.
Liquid handling robotics of various scales can automate steps of NGS sample preparation as well as NGS library preparation, providing the speed, throughput, and reproducibility necessary while minimizing the risk of human error. In critical applications, such as clinical diagnostics, this reliability can reduce the risk of low-quality data and the need to repeat preparation and sequencing on precious samples.
The importance of addressing sample throughput as a potential bottleneck in the workflow is increasing with the accelerating adoption of sequencing in clinical applications, such as cancer diagnostics where early and reliable diagnosis can impact long-term patient outcomes.
Genomic sequencing companies now offer both public and private NGS services for genetic testing. To remain cost-efficient and cost-effective, these companies need to maintain a high sample throughput, processing large numbers of NGS samples per day and keeping their sequencing systems busy churning out data.
The difference between NGS sample prep and library prep
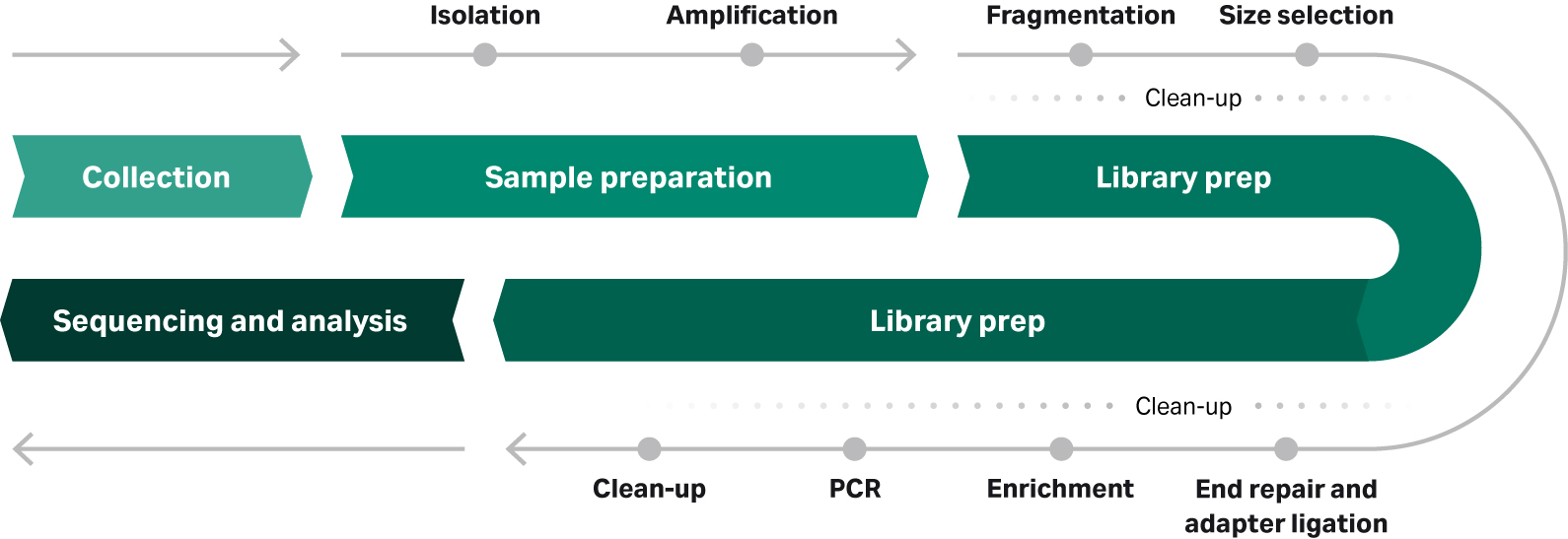
Between sample collection and sequencing in an NGS workflow, there are two key phases: sample preparation and library preparation, each containing a series of steps (Fig 1).
Fig 1: Typical NGS workflow from sample collection to sequencing and analysis. Sample preparation approach can vary considerably, dependent on the source, quantity, and quality of starting material.
NGS sample preparation involves sample-specific steps for extracting nucleic acids, such as genomic DNA from formalin-fixed, paraffin-embedded (FFPE) human tissue samples. These FFPE samples, for example, might require an extra step to repair damaged DNA to make them suitable for sequencing.
Sample preparation can also include an amplification step to increase the amount of nucleic acid fragments to a level sufficient for library preparation and sequencing instruments.
Library preparation takes the purified and amplified nucleic acid (DNA or RNA) from sample prep and processes them to be of optimum length by adding sequencing barcodes and adaptors. In some cases, a hybridization- or PCR-based enrichment step further optimizes the library for sequencing by enriching the specific regions of interest.
The different approaches to NGS sample preparation
There are various technologies and approaches enabling sample preparation for NGS. These range from traditional solution-based protocols, through solid-phase and membrane chemistries, to magnetic bead-based approaches.
Magnetic bead-based approaches are arguably a subcategory of solid-phase extraction, but there are numerous chemistries possible, and the workflow is distinct enough to consider as a separate type altogether.
Each of these approaches follows the three basic steps to DNA (or RNA) isolation:
- Lysis of cells in sample, varying from gentle detergents to aggressive homogenization.
- Removal of contaminants, including unwanted nucleic acids and denatured proteins.
- Recovery of nucleic acids into an appropriate buffer for any downstream application.
Table 1 lists several of these technologies and approaches to NGS sample prep.
Table 1: Overview of technologies and approaches to NGS sample preparation
| Type | Example method or chemistry | Key points |
| Solution-based | Phenol-chloroform (organic solvent) |
|
| Phi29-based amplification |
| |
| Solid-phase | Silica spin column |
|
| Magnetic bead-based | Silica coated magnetic beads |
|
| Oligo(dT) surface chemistry |
|
Solution-based sample preparation methods
Probably the most well-known and widely used solution-based method for nucleic acid isolation is phenol-chloroform DNA extraction. Although this method uses hazardous chemicals, when handled correctly it yields high-quality DNA at a low cost. The approach is common in research laboratories, where students and post-doctoral researchers perform phenol-chloroform extractions as part of daily routine.
However, the approach is not so amenable to high-throughput applications. It requires careful handling and could have phenol (an inhibitor) carryover. Also, although modern liquid handling robots are quite sophisticated, they cannot judge and achieve a balance between yield and purity on a sample-to-sample basis that becomes second nature to researchers.
As such, phenol-chloroform-based sample preparation is better suited for low-throughput, high-yield needs, rather than high-purity or throughput required by sequencing labs.
An alternative solution-based method that is more suited to automation involves Phi29 DNA polymerase-based DNA amplification. This approach faithfully amplifies limited samples, such as those from single cells, using a one-tube, one-temperature format.
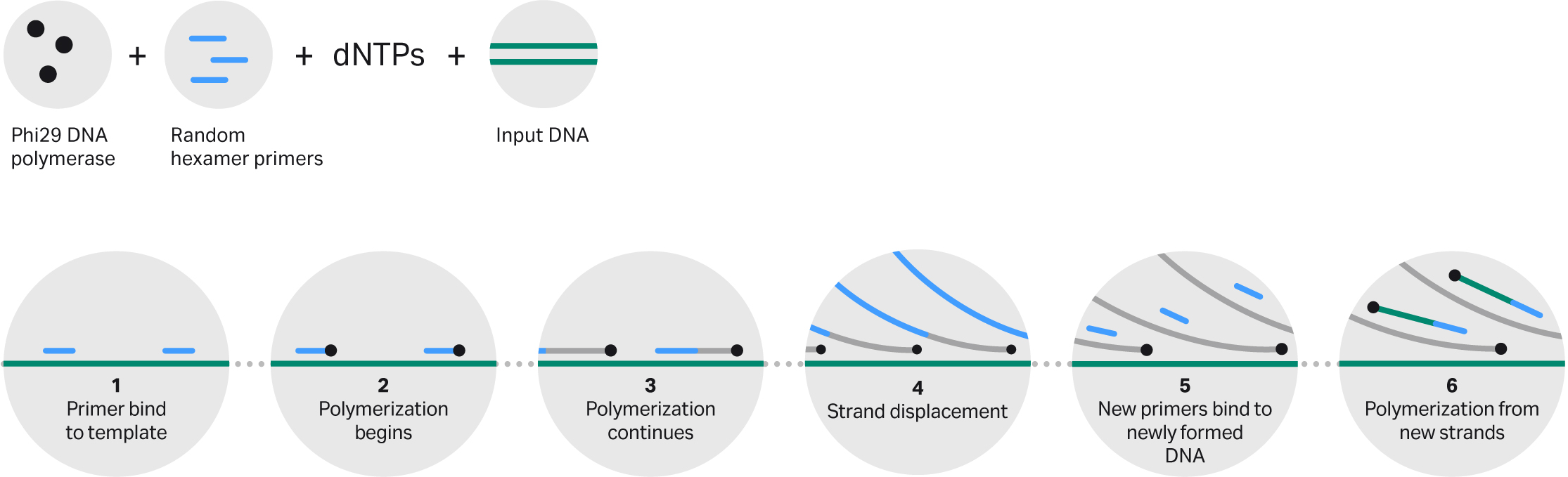
The high-fidelity Phi29 DNA polymerase is active at 30°C, does not require thermal cycling, and can produce micrograms of DNA from picograms of starting material. Phi29 relies on isothermal amplification, and can be used for both circular (or circularized) DNA template and linear DNA template (Fig 2).
Fig 2: Principle of Phi29 DNA polymerase-based amplification. Template DNA is primed at multiple locations with random hexamers. Phi29 DNA polymerase then extends primers, replicating template and displacing any downstream extended primers. Strand displacement and subsequent priming leads to an exponential increase of new template for isothermal amplification.
Commercial kits using Phi29 polymerase, such as illustra Genomiphi & TempliPhi DNA amplification kits, are designed for minimal hands-on time using simple automation-friendly protocols. These types of kits can also provide options for pre-dispensed reagents in single tubes, or multi-well plates, making them suitable for a range of throughput needs and liquid handling systems.
Solid-phase DNA isolation
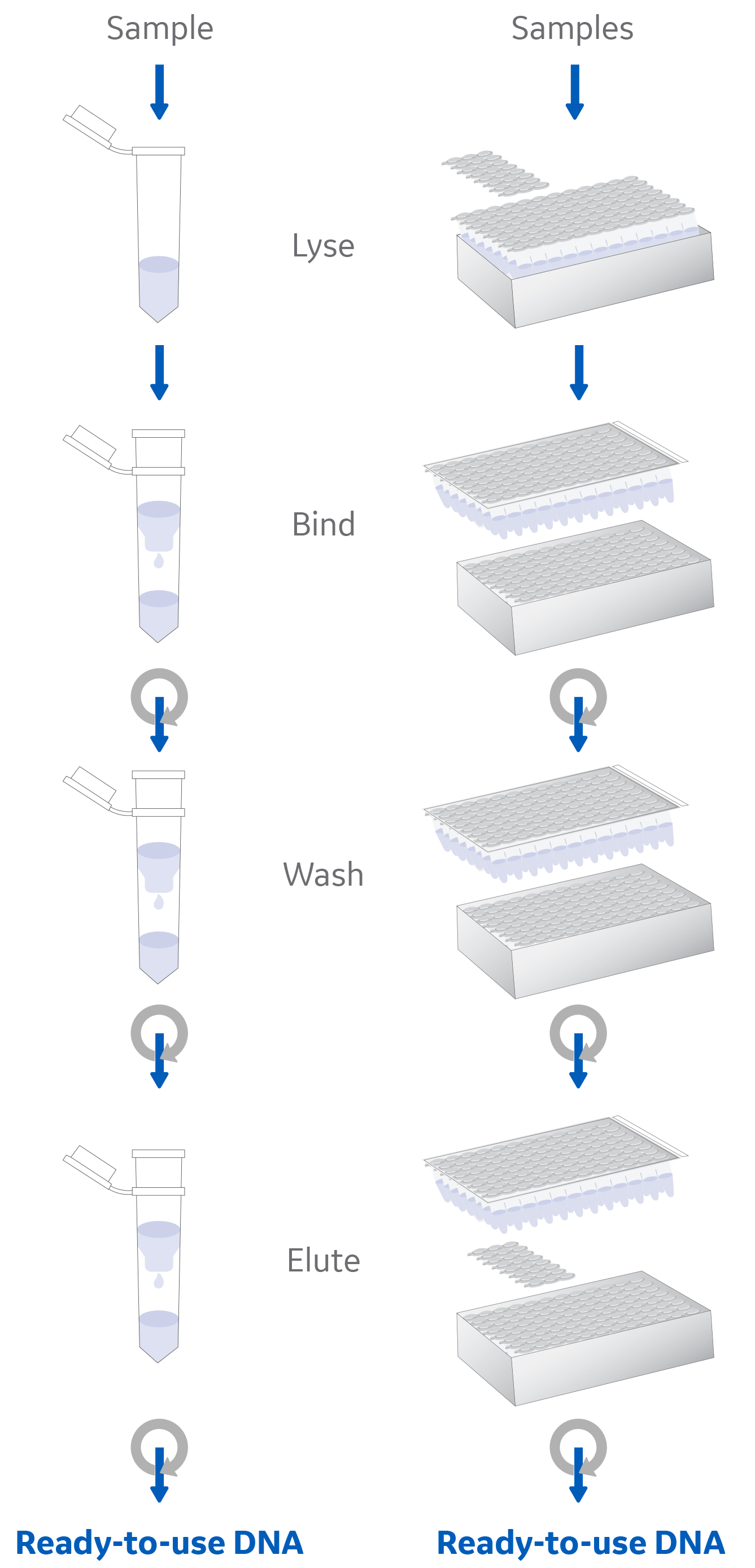
Solid-phase approaches to DNA extraction rely on the dependable chemistry between positively-charged silica and negatively-charged DNA. A typical protocol uses chaotropic salts to disrupt hydrogen bonds between strands, facilitating the adsorption of the nucleic acid phosphate residues to the silica membrane.
This silica-DNA association is predictable and easily modulated, enabling debris and potential contaminants to be washed away and purified DNA eluted with a high yield and purity (Fig 3).
Fig 3: Solid-phase nucleic acid extraction workflow using a silica membrane spin column.
Compared to the equivalent solution-based approach, phenol-chloroform extraction, silica spin-columns require fewer pipetting steps and all can be performed in the same vessel. These features make using silica spin-columns for NGS sample prep highly automatable with robotics and liquid handling systems, and so both suitable and in common use for high-throughput applications.
Magnetic beads-based nucleic acid isolation
Magnetic beads are a form of solid-phase nucleic acid isolation (DNA extraction and RNA extraction) technology, though there are also surface chemistries that enable capture of other substrates, such as proteins. These beads are composed of particles of iron oxides, such as magnetite, which give them superparamagnetic properties. That is, they exhibit magnetic properties only in the presence of an external magnetic field.
The protocols for using beads are scalable, simple to follow, and generally involve just three key steps that result in high-purity nucleic acid extractions (Fig 4), with no need for centrifugation or vacuum systems:
- Reversibly bind target molecules to magnetic beads added to any of a range of sample types.
- Apply a magnetic field to immobilize the beads while washing away the remainder of the sample.
- Adjust buffer conditions to release the purified target molecules.
Fig 4: Overview of magnetic bead-based nucleic acid isolation. (A) Target nucleic acid in the sample binds to beads based on buffer conditions. (B) Magnetic field immobilizes magnetic beads while unwanted debris and contaminants are washed away. (C) Beads release target nucleic acid by changing buffer conditions.
The range of surface chemistry options for magnetic beads covers a full spectrum of sample types, providing a level of versatility that is challenging to achieve with other approaches to NGS sample prep.
Silica-coated beads, for example, are well suited for the capture of high molecular weight genomic DNA from blood, tissue, and cell samples, even when the sample is scarce. For mRNA, oligo(dT)-coated beads provide the ability to purify directly from cell samples.
For applications where the sample is scarce, such as circulating cell-free DNA (cfDNA) sequencing, a combination of robust silica coatings and efficient buffer chemistry can provide the sensitivity needed to isolate the low molecular weight target molecules.
In each case, binding and releasing target molecules simply involves adjusting buffer conditions, making the workflow readily automatable with liquid handling robotics.
Magnetic bead-based DNA and RNA isolation, therefore, provides a means to avoid bottlenecks in sample preparation for a range of sample types while delivering reliable results and reproducibility in high-throughput environments.
These beads are straightforward to incorporate into sample preparation workflows, removing the need for hazardous solvents in the case of existing phenol-chloroform methods, and providing high binding capacity and scalability cost-effectively.
The magnetic bead-based sample preparation workflow is also simpler than that of solution-based methods or spin columns, with no requirement for centrifugation, vacuum systems, or indeed substantial experience or training to achieve high-purity DNA and RNA isolation.
Magnetic beads can also support NGS library prep in size selection and clean-up, supporting low- to high-throughput, consistency, and time and cost savings throughout the NGS workflow.
Closed vs open systems for NGS workflows
There are many commercial products for NGS sample prep. Most involve a degree of centrifugation, though some rely on vacuum manifolds. Some vendors offer these as part of automated “closed” systems, which provide both advantages and disadvantages to the user.
Compared to so-called “open” systems, where a user can select components from multiple manufacturers to develop a cost-effective protocol for their own purpose, closed systems limit the user to products from the same manufacturer or its partners. Manufacturers take this approach to ensure full compatibility between components and provide a level of reassurance that yield and quality will be consistent.
Large integrated systems can be coupled with other equipment to perform other NGS sample preparation steps, such as amplification. Although these integrated systems are well suited to high-throughput applications, they require a relatively large investment.
This level of investment might only be justifiable in certain situations, for example, high-throughput sequencing labs running clinical NGS panels. For these types of labs, having a single supplier means the entire workflow is already tried and tested, and a single point of contact for support minimizes the risk of delays during any troubleshooting.
Supporting high-throughput sequencing providers
We have many years of magnetic beads, with an R&D team experienced in developing products that help maximize operational efficiency in high-throughput environments.
In addition to a range of existing nucleic acid isolation kits designed for use with a full spectrum of sample types including cultured cells, tissue, blood, plant cells, and bacteria, the team supports customers with customized solutions. For more information, contact the Life Sciences Support team.
- Sera-Xtracta Cell-Free DNA Kit
- Sera-Xtracta Genomic-DNA Kit
- Sera-Xtracta Virus/Pathogen DNA Kit
- SeraSil-Mag silica coated superparamagnetic beads
- Sera-Mag Oligo(dT)-Coated Magnetic Particles
- Sera-Mag SpeedBeads and Sera-Mag Carboxylate-Modified Magnetic Particles
- illustra TempliPhi Amplification kits