Explore the mRNA universe with Cytiva. In this series, our scientists share their journey and insights on mRNA synthesis for research purposes. And they share their advice on how to scale. Through peer-to-peer interactions, we continue to encourage the spirit of collaboration developed at a time when new modalities are being used as therapies for the first time.
“When it comes to mRNA, many manufacturing investments are being made while still early in the process development workflow. So essentially, we are translating methods from the milliliter scale to the liter scale. While doing this, I realized that a lot of the decisions made on mRNA process will have a great impact on equipment setup, batch cost, and throughput.” — Katarina Stenklo, Enterprise Solutions, Cytiva
Part 1 – Introduction to mRNA manufacturing
Over the past two years, the emergence and worldwide spread of the SARS-CoV-2 virus, the cause of coronavirus disease 2019 (COVID-19), has changed the course of scientific research around messenger RNA (mRNA). In fact, the development of mRNA vaccines is undoubtably one of the most significant scientific breakthroughs that has contributed to bringing the world out of the pandemic.
On its own, “naked” mRNA degrades rapidly after injection, a fact that has led scientists to pursue the successful and currently widely used approach of encapsulating mRNA into lipid nanoparticles (LNPs). LNPs act to protect and deliver mRNA to target cells and support cell uptake by endocytosis. Once in the cytoplasm, mRNA is released from the LNP and is translated into a protein. This has led to intensified research within the mRNA–LNP field, and mRNA-based therapeutics are now viewed as key solutions for tackling some of the most difficult and pressing clinical challenges such as cancer, genetic, and infectious diseases.
This series aims to guide you in the practical aspects of setting up an mRNA research laboratory, addressing methods, equipment, reagents, and workflow. Even if your current aim is not process development with the goal of scaling up, it is nevertheless beneficial to consider the feasibility and ease of scaling up. This will in turn save you precious time in the future. Please note that not all methods described here are scalable.
Structural features of mRNA

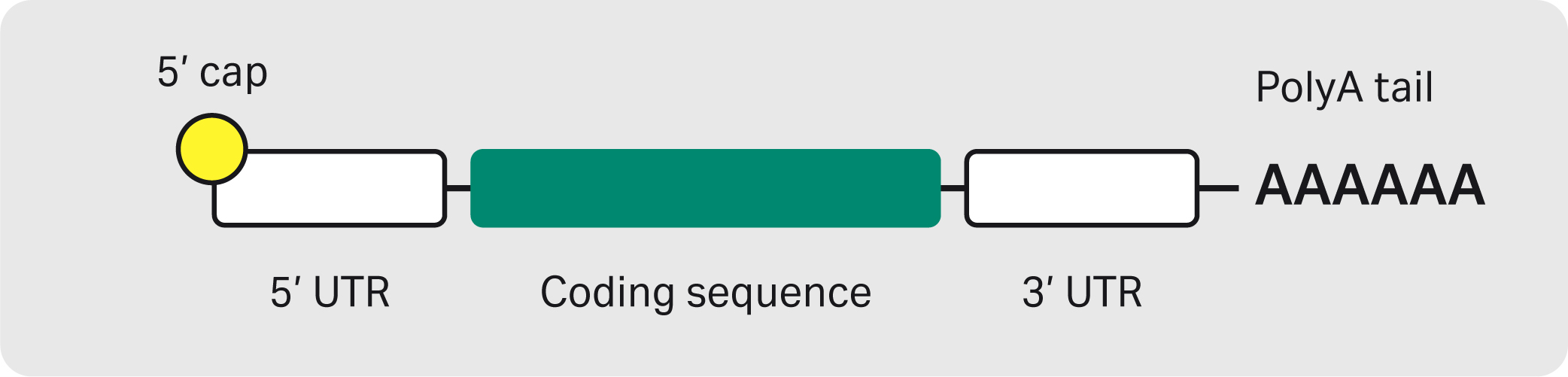
In almost all eukaryotes, modifications are made at both ends of the nascent mRNA, a 5´ cap and a 3’ polyadenylated (polyA) tail. The 5’ cap is multifunctional and is involved in mRNA maturation, nucleocytoplasmic transport, and protection from 5‘ exonuclease digestion. The polyA tail, a chain of adenine nucleotides at the 3’ end, increases stability of the molecule and appears to play a role in mRNA quality control. In addition to these modifications, a mature mRNA also consists of a 5’ untranslated region (UTR), a coding sequence, and a 3’ UTR (Fig 1). UTRs are known to play a role in the post-transcriptional regulation of gene expression and are a part of the “gene” or DNA template design in the in vitro transcription (IVT) process. During mRNA manufacturing, capping of the transcribed mRNA can be done in different ways, for example in the IVT chamber directly (co-capping) or as an additional enzymatic step downstream of IVT. The polyA tail is usually included in the DNA template.
Translation happens outside the nucleus, where the mRNA serves as a template to construct a protein from amino acids. The 5’ cap tags the mRNA as self-RNA to the innate immune system and initiates ribosomal binding. Together with the 5’ cap, the polyA tail promotes translation to protein. UTRs modulate translation efficiency, and the coding sequence contains your gene of interest. The polyA tail promotes mRNA stability (3’ exonuclease protection is one such mechanism) and participates in terminating transcription.
Even a single change (e.g., strand break) in the mRNA strand can halt translation, making it vital to monitor the integrity of the molecule during the process.
Fig 1. The structure of a mRNA molecule.
This series describes several analytical assays, including PCR, chromatography, and filtration methods. For a more comprehensive list, please refer to a table compiled by Schoenmaker and co-workers (1).
RNase-free considerations
Ribonucleases (RNases) are ubiquitous and present the biggest risk when working with RNA due to their ability to rapidly degrade RNA. Hence, it is critical to emphasize the importance of maintaining an environment free from RNases. Besides being potentially present in reagents, tips, tubes, bottles, and surfaces, RNases are also found in bodily fluids (e.g., skin oil and perspiration). You can minimize RNase contamination by:
- Using exclusively RNase-free reagents, solutions, and equipment
- Wearing protection such as a face mask, gloves, and lab coat
- Changing gloves often
- Decontaminating surfaces and pipettes diligently
- Dedicating equipment and material and labeling them “RNase-free” or “for RNA work only”
- Using certified RNase-free consumables such as pipette tips with barriers and tubes
- Ideally creating a separate mRNA lab or an area with equipment and material specifically dedicated for RNA work
In addition, it has been suggested that autoclaving or baking glassware for at least two hours at 240oC can destroy RNase activity. Unfortunately, these enzymes are very robust and will still retain partial RNase activity. Although diethyl pyrocarbonate (DEPC) has been used to inactivate RNases, leftover traces or byproducts may inhibit subsequent enzymatic reactions. Furthermore, DEPC is incompatible with Tris-based buffers such as IVT buffer, which contains Tris-HCl.
RNases cannot be easily inactivated by ethanol, ethylenediaminetetraacetic acid (EDTA), or other metal chelators. Highly concentrated sodium hydroxide (NaOH) is effective against RNases, but soaking is time-consuming.
For solutions (e.g., buffers) prepared in house, it is highly recommended to test with an RNase activity test kit such as RNaseAlert® kit from Integrated DNA Technologies (IDT). The result can be read visually for qualitative assessment of contamination or quantified using fluorometry. IDT also offers an anti-RNase reagent, Nuclease Decontamination Solution, that can easily be applied on and around your workspace to irreversibly inactivate RNases and reduce contamination risk. Moreover, it can be applied to plastic surfaces, which are difficult to sterilize.
“RNases are extremely resistant to most chemical and biological conditions. For instance, RNase A (Thermo Fisher Scientific) tolerates both boiling and autoclaving. So far, there is no RNase elimination method that is compliant with biopharmaceutical production. That means prevention and monitoring are the best ways to control RNase contamination.” — Wangshu Jiang, R&D Scientist, Uppsala, Sweden
In part 2, we will look at challenges and strategies related to the DNA template process.
References:
- Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int J Pharm. 2021;601:120586.