Explore the mRNA universe with Cytiva. In this series, our scientists share their journey and insights on mRNA synthesis for research purposes. And they share their advice on how to scale. Through peer-to-peer interactions, we continue to encourage the spirit of collaboration developed at a time when new modalities are being used as therapies for the first time.
Part 5 – LNP formulation
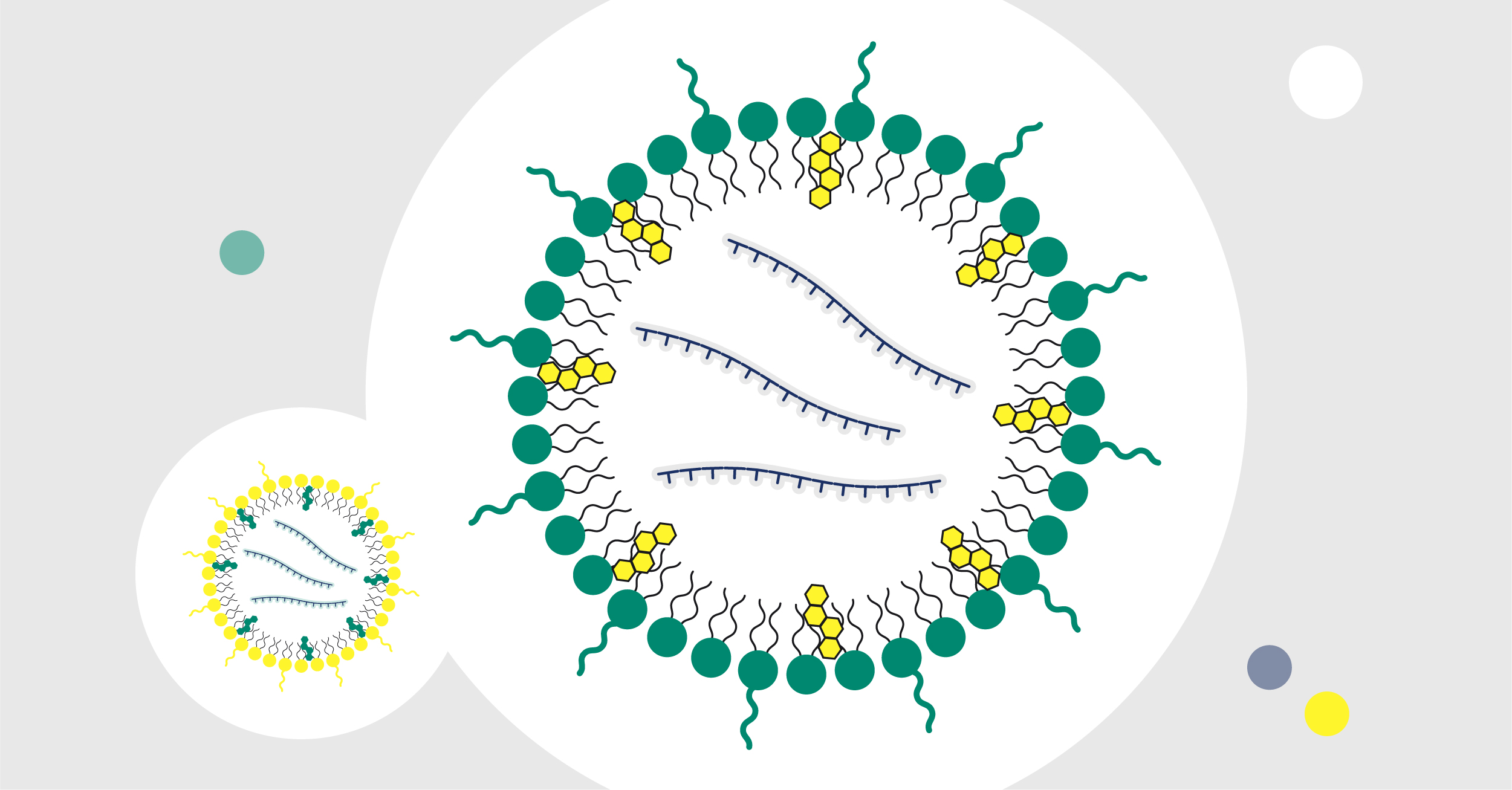
Microfluidic mixing is one of the most effective means to formulate lipid nanoparticles (LNPs) because of its scalability and reproducibility. An aqueous buffer containing mRNAs and an organic lipid mixture are rapidly mixed in a controlled and non-turbulent manner. This step facilitates self-assembly into stable nanostructures such that mRNAs are encapsulated in the interior core through interactions with ionizable lipids, cholesterol, helper lipids, and stabilizers. This protective structure allows the mRNA to evade degradation by endosomes and increases stability in physiological fluid. It’s important to consider how the combination of cationic or ionizable lipids and the lipid composition (i.e., the ratio of helper lipids and surfactants) impact mRNA encapsulation.
After mRNA is encapsulated, the LNPs are diluted with a suitable aqueous buffer to reduce the organic solvent content. Nanoparticle size and polydispersity index (PDI), which gives the size distribution of nanoparticles, can be determined by using a dynamic light scattering bioanalyzer. A high PDI indicates a broad or multimodal size distribution. Hence, the lower the PDI, the better. Encapsulation efficiency can be measured using fluorescence-based, RNA-specific assays.
Challenge
- Need for reliable and user-friendly encapsulation systems
- Need for a ready-to-use lipid mixture that has the right combination of lipids and lipid composition
Strategy
- Precision Nanosystems offers a range of systems and reagents based on a microfluidic mixing technique, resulting in reproducible and high-quality LNPs.
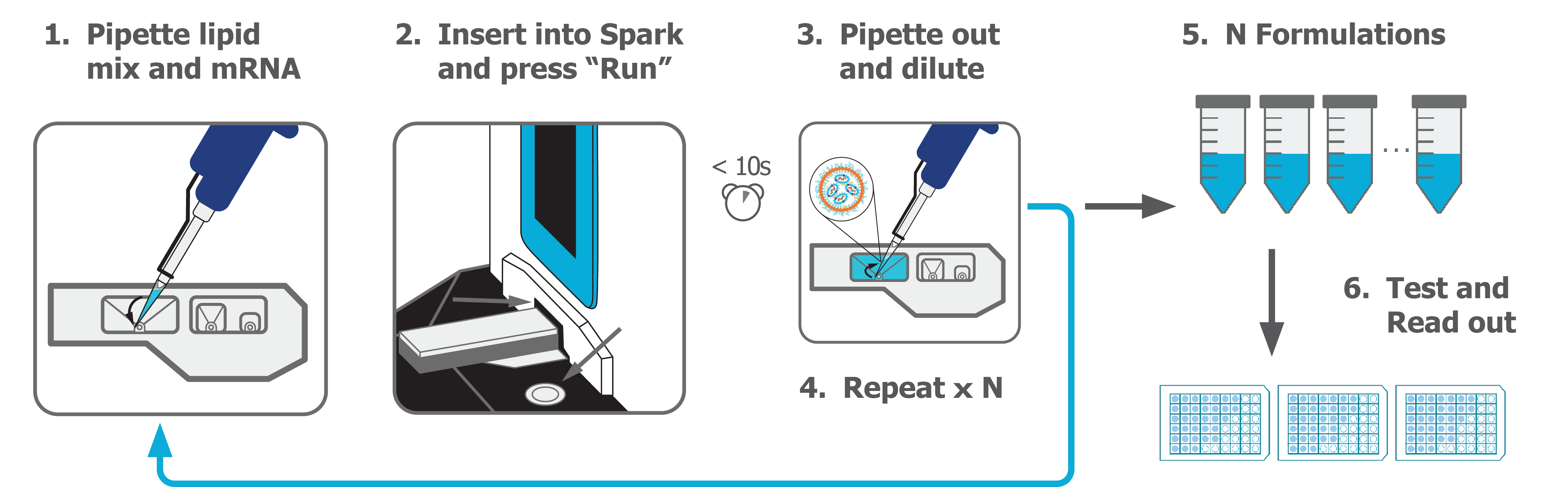
NanoAssemblr® Spark™ is the smallest system that formulates between 25–250 μL of LNP, encapsulating mRNA on the microgram scale (Fig 1). The single-use NxGen™ microfluidic mixer cartridge minimizes cross-contamination, and simple one-button operation reduces user variability. The petite Spark™ system can easily be placed on the bench-top or in a sterile biosafety hood. Since formulation takes barely 10 s per run, LNPs can be made on-demand for immediate use.
Fig 1. Precision Nanosystems NanoAssemblr® Spark™ system (top) and workflow (bottom).
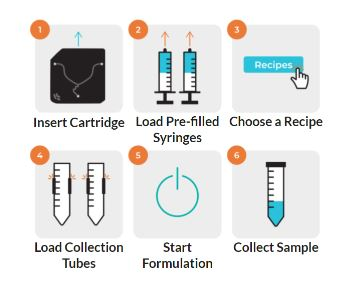
NanoAssemblr® Ignite™ is the larger sibling of the Spark™ system, capable of formulating 1–20 mL. By adjusting key parameters such as flow rate ratio, total flow rate, and dilution, lipid composition is optimized to obtain the desired LNP size and structure Formulation times are short (< 1 min/run, total flow rate up to 20 mL/min) allowing formulations to be made on-demand. The setup is easy with a touch screen interface. Recipes are reproducible and can be stored. The optional in-line dilution via the choice of appropriate single-use NxGen™ cartridges enhances flexibility.
Fig 2. Precision Nanosystems NanoAssemblr® Ignite™ system (left) and workflow (right).
- GenVoy-Ionizable Lipid Mix (GenVoy-ILM™) is a pre-optimized Research Use Only lipid mixture designed for encapsulation and can be used at various stages of drug development from discovery to late preclinical, effectively delivering mRNA to cells with high efficiency and low toxicity. It is comprised of lipid components at defined ratios and is optimized for use with the NanoAssemblr® systems. This lipid mixture circumvents formulation-related challenges, allowing a quick start to setting up an mRNA-LNP laboratory.
LNP polishing
This step exchanges the buffer and removes excess lipids and non-encapsulated mRNA to further increase the purity. This fast and relatively inexpensive process can be readily executed in the laboratory by centrifugal ultrafiltration. The primary basis for separation is molecular size. Molecules larger than the membrane pores will be retained, but not bound, at the surface of the membrane and concentrated during the ultrafiltration process. The retention properties of ultrafiltration membranes are expressed as molecular weight cut-off (MWCO), which is based on the ability to retain > 90% of a solute of a known molecular weight (in kD). Different membranes are available, and it's important to choose the ones with low nonspecific biomolecule binding that can provide > 90% recovery. At larger scales, centrifugal ultrafiltration will be replaced with tangential flow filtration systems.
Challenge
- Need for high-quality centrifugal ultrafiltration devices with a wide range of MWCOs and suitable membrane
Strategy
- Centrifugal ultrafiltration devices from Pall Corporation allows polishing of LNPs for volumes of < 50 µL to 60 mL. For individual samples, there are various sizes (Nanosep®, Microsep™, Macrosep® and Jumbosep™) to choose from. For automation purposes, AcroPrep™ plates come as 24-, 96-, and 384-well filter plates, suitable for volumes of 80 µL to 7 mL. Omega™ (modified polyethersulfone) ultrafiltration membrane provides high flow rates and low protein binding and is available in a variety of MWCOs.
Sterile filtration of mRNA-LNPs
This final sterile filtration step removes potential microbial contaminants. The same filters (
Summary
Start your mRNA research laboratory with confidence when you leverage the combined expertise and long history within the Danaher family. Cytiva, Integrated DNA Technologies, Aldevron, Precision NanoSystems, and Pall Corporation are here for you every step of the way, with a comprehensive range of quality reagents and systems across the mRNA production workflow.