Explore the mRNA universe with Cytiva. In this series, our scientists share their journey and insights on mRNA synthesis for research purposes. And they share their advice on how to scale. Through peer-to-peer interactions, we continue to encourage the spirit of collaboration developed at a time when new modalities are being used as therapies for the first time.
Part 4: mRNA purification
“One of the biggest challenges you’ll face is maintaining the stability of the mRNA while maximizing yield and purity. Keeping the reaction and all equipment in RNAse-free conditions is a must. But don’t assume it’s RNase if you’re having problems – the type of buffer and salt concentration, as well as time and temperature, also affect the stability of the mRNA during purification.”
Susanna Lindman, Principal Scientist at Cytiva R&D, Uppsala, Sweden
Impurities from in vitro transcription (IVT) – including DNA template, enzymes, NTPs, salts, double-stranded (ds) RNA, and truncated mRNA – can induce undesired immune responses and negatively impact translation efficiency. For these reasons, the mRNA must be purified. There are several options to purify mRNA: lithium chloride (LiCl) precipitation; oligo deoxythymidine (dT) magnetic particles; and chromatography. When designing a purification process, consider scalability and suitability for a manufacturing environment. Read on to discover the pros and cons of each option.
DNA removal by DNase I digestion followed by LiCl precipitation is a common laboratory-scale purification method for mRNA. LiCl precipitation uses elevated concentrations of lithium cations to selectively precipitate mRNA, leaving behind impurities. The precipitated mRNA is pelleted, and the supernatant is discarded. Although mRNA is specifically separated from DNA, DNase I treatment is still recommended for complete DNA removal. EDTA can be used to inactivate DNase I after digestion. A major disadvantage is that undesired aberrant mRNAs such as dsRNA and truncated mRNA are still present.
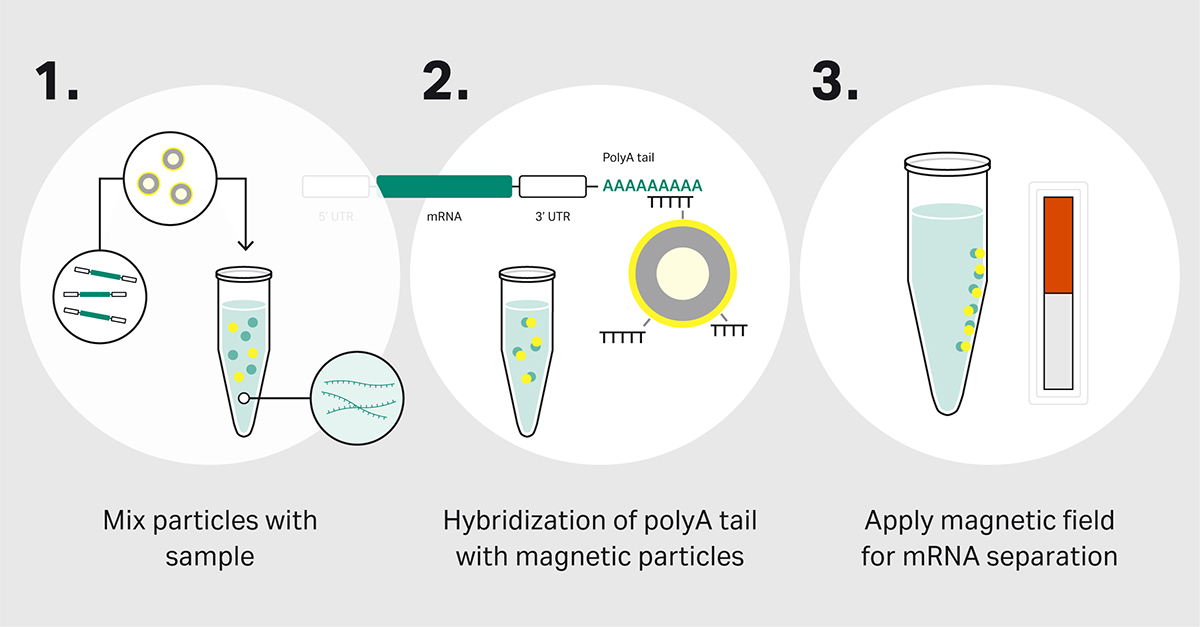
Separation via oligo(dT) magnetic particles is based on specific hybridization to the polyA tails. The process is simple (Fig 1). When the particles are added to the sample, the polyA tails hybridize to the oligo(dT) ligands on the surface of particles. When an external magnetic field is applied, the beads with the bound mRNA are attracted and immobilized to the outer edge of the tube. The contaminants are removed during the washing steps. Next, the addition of an elution buffer releases mRNAs from the immobilized beads as a purified sample. Finally, the magnetic field is removed, and the beads are released, ready for re-use.
Fig 1. mRNA capture with oligo(dT) magnetic particles.
With this method, there’s no need for centrifugation or vacuum, eliminating stress or shear force. In addition, the process is amenable to automation in 24-, 96-, and 384-well plates. Like LiCl precipitation, DNase I treatment is optional but recommended. Untailed mRNAs are removed, but other forms of aberrant mRNAs with polyA tails remain. Neither LiCl precipitation nor oligo(dT) magnetic particle separation is scalable.
Chromatography, a purification process routinely used in the pharmaceutical industry, is gradually replacing traditional precipitation methods. This is true because chromatography offers higher reproducibility, recovery, and purity as well as better scalability. Another advantage is that chromatography can effectively remove DNA template, aberrant RNA, residuals, and impurities. A variety of techniques are available, including oligo(dT) affinity, anion exchange, hydrophobic interaction, and multimodal chromatography.
Oligo(dT) affinity chromatography captures mRNA by binding to the polyA tails via oligo(dT)-tagged beads. DNA, NTPs, enzymes, buffer components, and any other impurities without polyA tails are washed away. The downside is that aberrant RNAs aren’t removed, which is why this step is usually followed by another chromatography step. Anion exchange chromatography (AEX) is an attractive option for polishing, because it provides good separation of single-stranded mRNA from immunogenic DNA, RNA-DNA hybrids, dsRNA, and other aberrant RNAs.
AEX is based on the reversible interaction between the negatively charged sugar-phosphate backbone of mRNA and the positively charged chromatography resin. An alternative is hydrophobic interaction chromatography, which is based on the reversible reaction between mRNA and the hydrophobic ligand of the resin. Core bead chromatography, a form of multimodal (mixed-mode) chromatography, is another effective technique to remove low molecular weight impurities. It combines the power of size exclusion and binding separation such that small impurities are trapped inside the beads and the large mRNA product flows through.
After purification, the mRNA can be quantified with either fluorescence-based, RNA-specific assays or a spectrophotometer. An absorbance ratio (A260/A280) less than 2.0 may indicate the presence of residual DNA or protein, and the mRNA product may not be suitable for downstream applications unless further purified. High performance liquid chromatography (HPLC), enzyme-linked immunosorbent assay (ELISA), or dot blotting can be used to check for unwanted truncated or dsRNA. It’s important to prefilter mRNA prior to an HPLC run to remove particulates that can damage columns.
The stock concentration for encapsulation is 1 mg/mL. The concentration of mRNA may need to be adjusted after purification, by diluting in a suitable aqueous buffer. To reduce the risk of mRNA degradation, work quickly and wear gloves. If there’s time between the steps, consider keeping the mRNA cold.
Challenges
- Choosing the optimal combination of methods to remove impurities, especially aberrant mRNA
- Choosing the analysis method for aberrant RNA
Strategies
Choose mRNA capture, polishing, and analysis products from Cytiva:
- Capture mRNA using Sera-Mag™ Oligo(dT) magnetic particles
- Perform microscale purification by converting ÄKTA pure™ 25 chromatography system using a Micro kit
- Simplify the workflow with HPLC-certified regenerated cellulose (RC) filters, as they’re compatible with many solvents to minimize extractables
- Perform dot blotting with Nytran™ SuPerCharge (SPC) nylon membrane and Cy™3/Cy™5 secondary antibodies
Sterile filtration of mRNA
Prior to encapsulation, perform a sterile filtration step to remove potential microbial contaminants.
Challenge
- Need for high-quality 0.2 µm filters
Strategy
- 0.2 µm pre-sterilized Acrodisc® syringe filters from Pall Corporation equipped with Supor® or Fluorodyne® membranes are good for filtering fluids up to 150 mL with a holdup volume as low as 60 µL and are available in 13, 25 and 32 mm sizes. Fluorodyne® filters incorporate a high-capacity asymmetric polyethersulfone (PES) prefilter layer over a polyvinylidene fluoride membrane layer, while Supor® filters have a PES membrane with broad pH compatibility. Sterilization by gamma irradiation prevents the contamination risk that can result from ethylene oxide sterilization
“Every additional step reduces the total process yield and increases the amount of consumables used and therefore the total process cost.” Katarina Stenklo, Enterprise Solutions, Cytiva
In part 5, we will look at challenges and strategies related to the LNP formulation process.