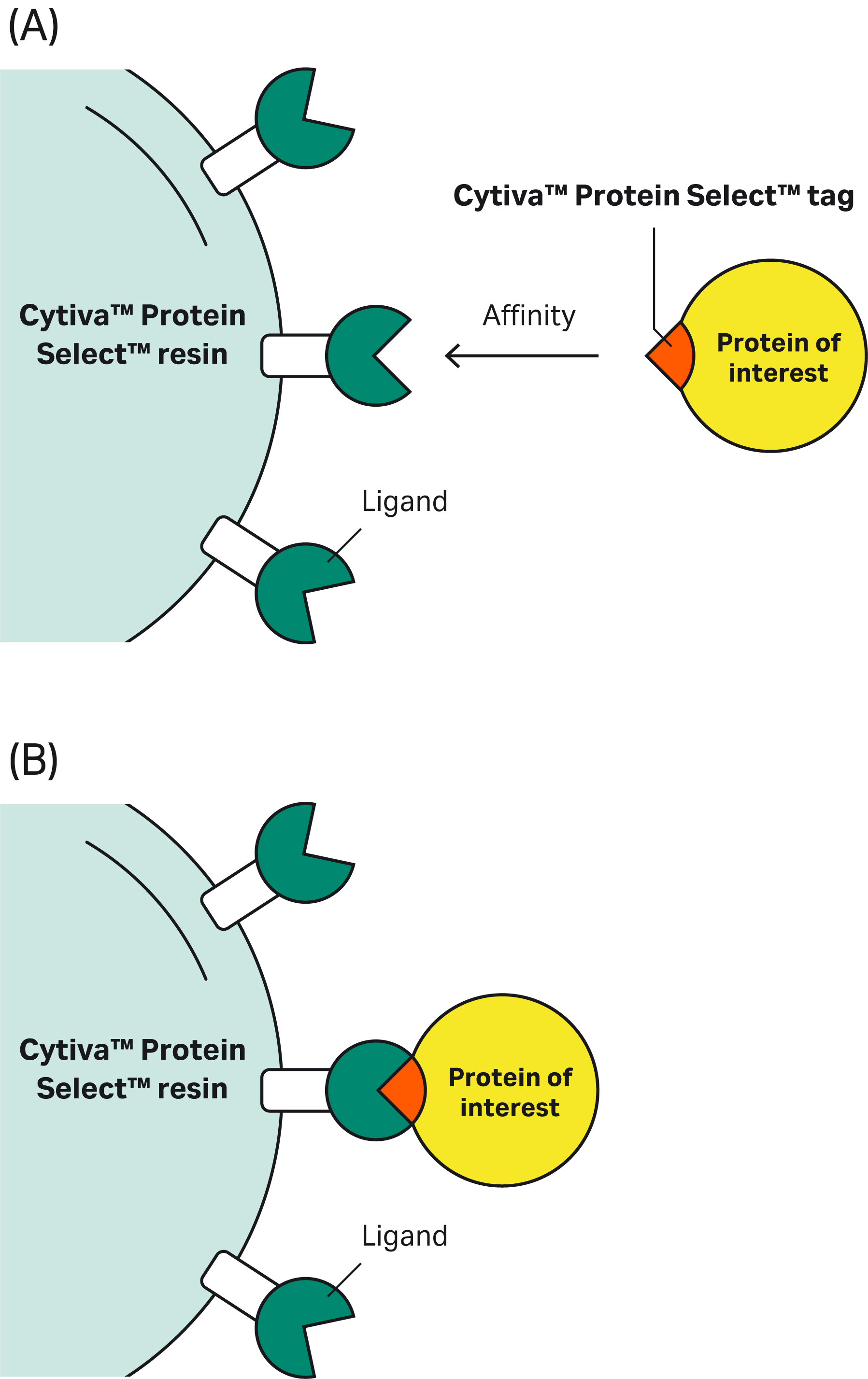
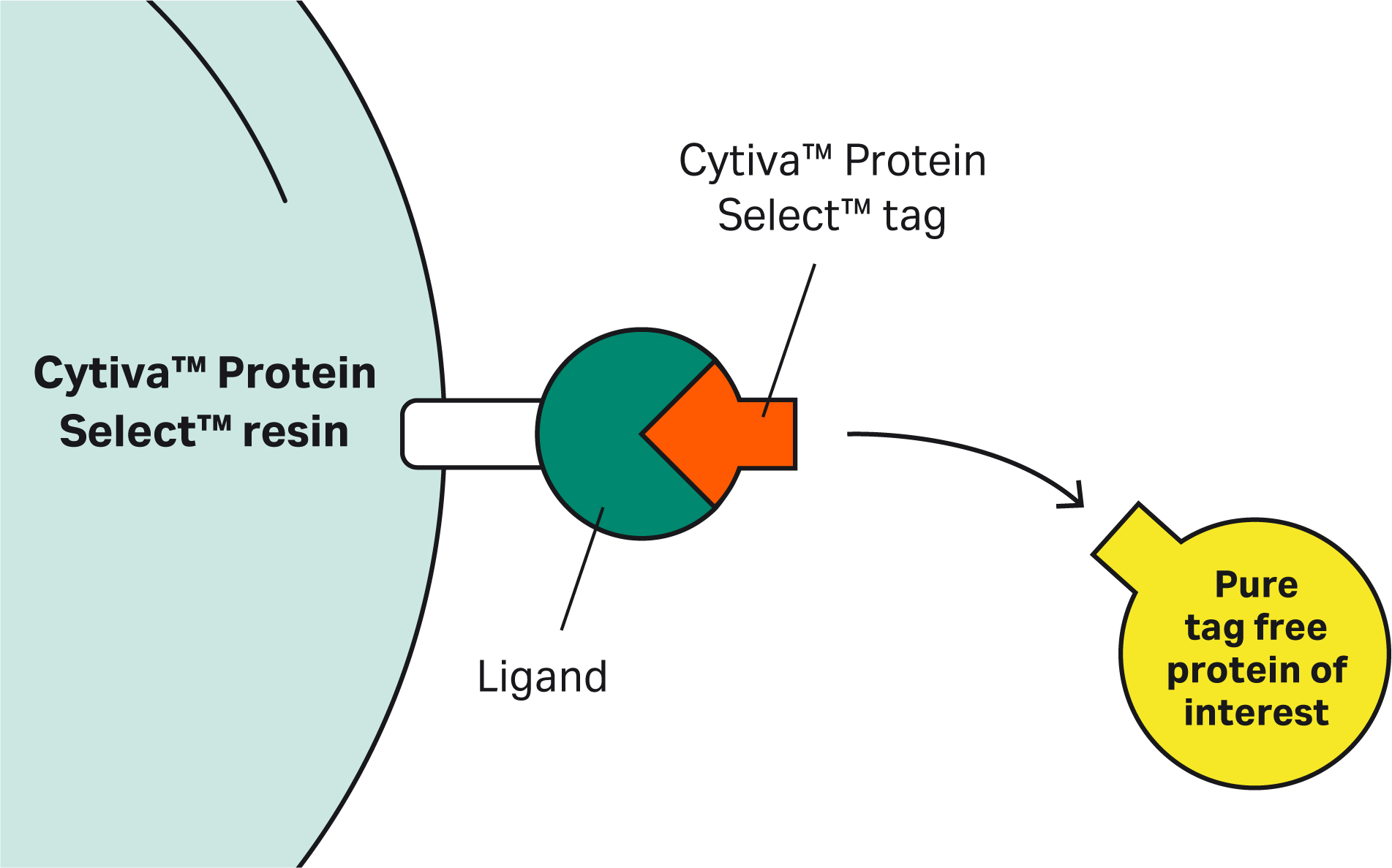
Answers to a range of questions covering the mechanism, application, capabilities, and potential use cases of the resin and tag for purifying recombinant proteins
With questions covering a range of topics from principles to protocols, we’ve heard from scientists who are interested in learning more about using Cytiva™ Protein Select™ technology to purify proteins that lack an affinity binding partner. Their questions and our answers are collected here.
Want to discuss the Cytiva™ Protein Select™ system for purifying your protein? Contact us!
- I want to discuss research applications
- I want to discuss process development/bioprocessing applications
Additives
| Question | Answer | |
|---|---|---|
| 1 | Are there any reagents that will prevent purification if they are present in the sample? | High concentrations of denaturing substances will prevent purification. Other than that, we’ve tested many common buffers, salts, and excipients, including EDTA, DTT, and divalent metal ions, and we haven’t seen purification problems with them. |
| 2 | Is it possible to purify samples that contain detergents for membrane proteins? | Yes. n-Decyl-β-D-maltoside (DM) can be used in buffer and sample. Lauryldimethylamine oxide (LDAO) can also be used, although its use has coincided with some decrease in target protein yield. However, we have not yet purified a membrane protein ourselves. We used a CelLytic Express lysis kit (Sigma-Aldrich) for extraction of target proteins produced intracellularly in E. coli, and subsequent purification was successful. The kit does not say exactly what the detergents are but mentions a blend of detergent and enzymes. |
| 3 | One of the proteins I want to express with the system would need to be purified totally under denaturing conditions. Can the whole purification process (including binding, cleavage, etc.) be done in 4-8 M urea? | No. Purification cannot be run under denaturing conditions. However, a low concentration of urea (such as 1 M) can be present in the sample and buffer during purification. |
| 4 | Our protein has a tendency to oligomerize via S-S bridges. How does Cytiva™ Protein Select™ technology deal with this phenomenon? Is adding antioxidants to the buffer compatible with the technology? | For the excipients, you can use reducing agents; antioxidants and ascorbate would work. |
| 5 | We are working with insoluble protein and therefore using 8 M urea. Would this work with Cytiva™ Protein Select™ resin? | No. 8 M urea in the binding buffer will not work, because the tag needs to bind to the ligand on the resin and the complex needs to fold, which will be difficult to achieve in the presence of 8 M urea. Low concentrations of urea, such as 1 M, can be used. |
| 6 | Is the resin compatible with buffers containing amino acids, for example 50 mM to 1 M arginine (to prevent aggregation) or 10 mM histidine buffer? | Probably. We haven’t tested this, but based on what we know, the resin should be compatible with amino acid buffers. A pH between 6 and 9 and ionic strength corresponding to 0-1 M NaCl works. |
| 7 | Can the eluate from an immobilized metal chelate affinity chromatography (IMAC) purification be directly applied to the Cytiva™ Protein Select™ column (i.e., without removing imidazole)? | Yes. You can apply samples containing imidazole to Cytiva™ Protein Select™ resin. |
| 8 | Is it possible to use the tag system in the presence of reducing agents? | Yes, you can use reducing agents (e.g., DTT) during purification. |
Binding capacity
| Question | Answer | |
|---|---|---|
| 1 | What is the binding capacity of Cytiva™ Protein Select™ resin? | Because the tagged protein cleaves once bound to the resin, it is not possible to measure the traditional dynamic binding capacity (QB10) using UV signal. Yield will depend on protein design and cleavage time (hold time). We have observed yields (the amount of cleaved target protein in the eluate) up to 20 mg protein per mL resin. |
| 2 | How do I measure binding capacity or recovery? | You can add a detection tag or epitope tag and use that to analyze tagged protein in starting material, flowthrough, wash, and CIP fractions during operation. You can analyze eluted tagless protein by methods such as analytical size exclusion chromatography. |
CIP/regeneration
| Question | Answer | |
|---|---|---|
| 1 | How do I regenerate the resin? | For effective regeneration and cleaning-in-place (CIP) between purification cycles, we recommend using one of the following solutions with a contact time of 15 min: • 6 M urea with 50 mM NaOH • 4 M urea with 100 mM NaOH, or • 4 M guanidine hydrochloride with 100 mM NaOH. Alternatively, you may be able to use 30% isopropanol with 100 mM NaOH depending on sample composition. |
| 2 | Is it okay to regenerate the resin only with denaturants or NaOH? | No. Chaotropic agents (urea or guanidine-HCl) alone or NaOH alone will NOT effectively regenerate/CIP the resin. To effectively regenerate/CIP the resin between purification cycles, one needs a combination of chaotropic agents (urea or guanidine-HCl) and NaOH. |
| 3 | For the sanitization of the resin, it is indicated to use 4 M urea in 0.1 M NaOH. What is the stability of urea at this pH (~13)? | Urea solutions should always be freshly prepared and used, as solutions of urea may develop cyanate ions upon standing, especially at higher temperature. For small-scale purifications, we prepare aliquots of the regeneration solutions and store them at –20°C. |
| 4 | Can you strip the tag and reuse the resin? | Yes. Regeneration and CIP is possible with a combination of chaotropic agents (urea or guanidine-HCl) and NaOH. |
| 5 | How many times can the resin be reused? |
We tested 20 purification cycles of E. coli lysates with a CIP/regeneration solution of 4 M urea and 100 mM NaOH, and measured yield (amount of eluted protein) for each cycle. After 20 cycles we obtained 80% of the initial yield. We also tested a CIP/regeneration solution of 4 M guanidine HCl and 100 mM NaOH. The resin retained more than 65% of its initial yield after 50 purification cycles with E. coli lysates, and more than 86% of its initial yield after 20 purification cycles using a HEK293 feed. Different proteins were used in the CIP/regeneration experiments. |
Cleavage mechanism
| Question | Answer | |
|---|---|---|
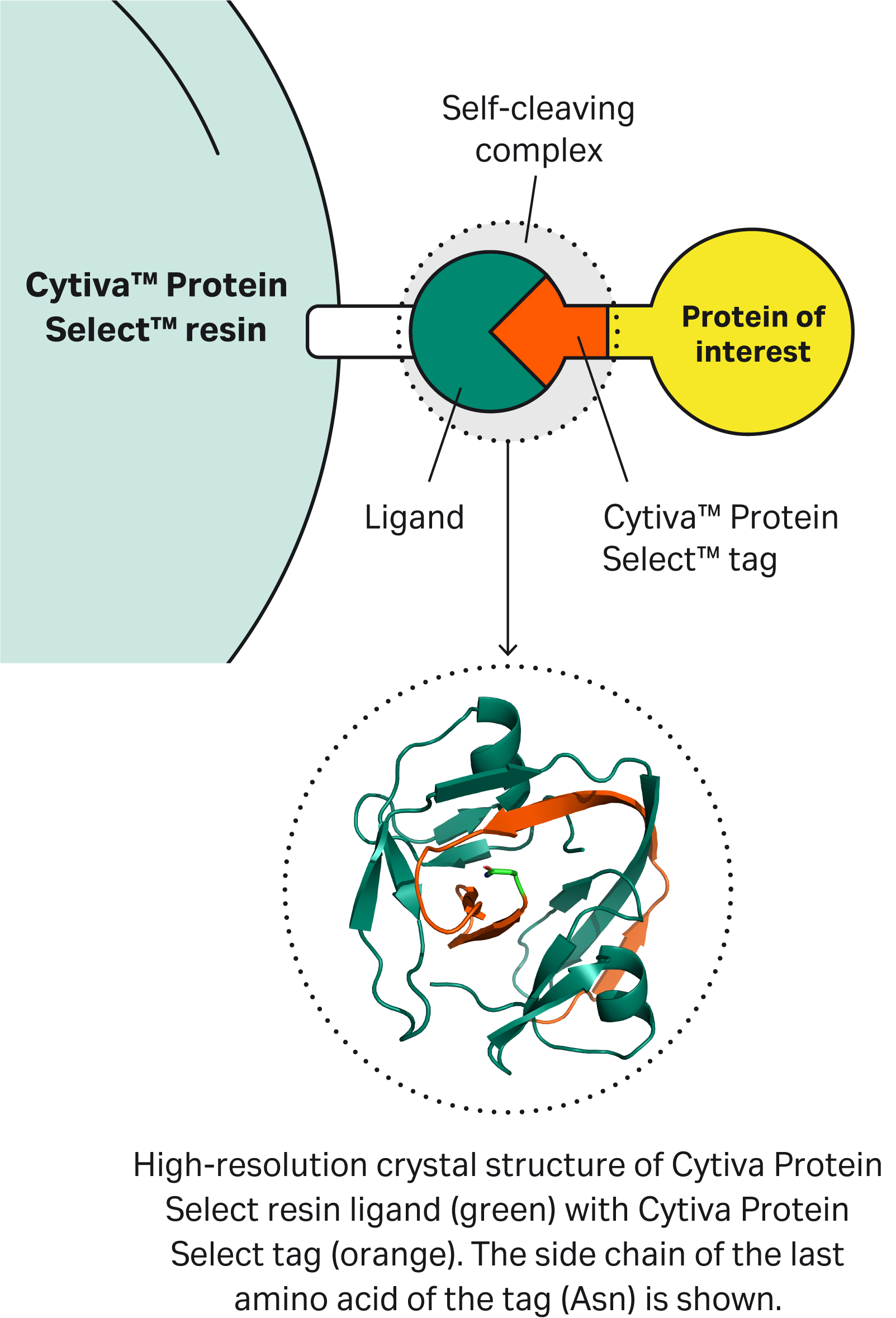
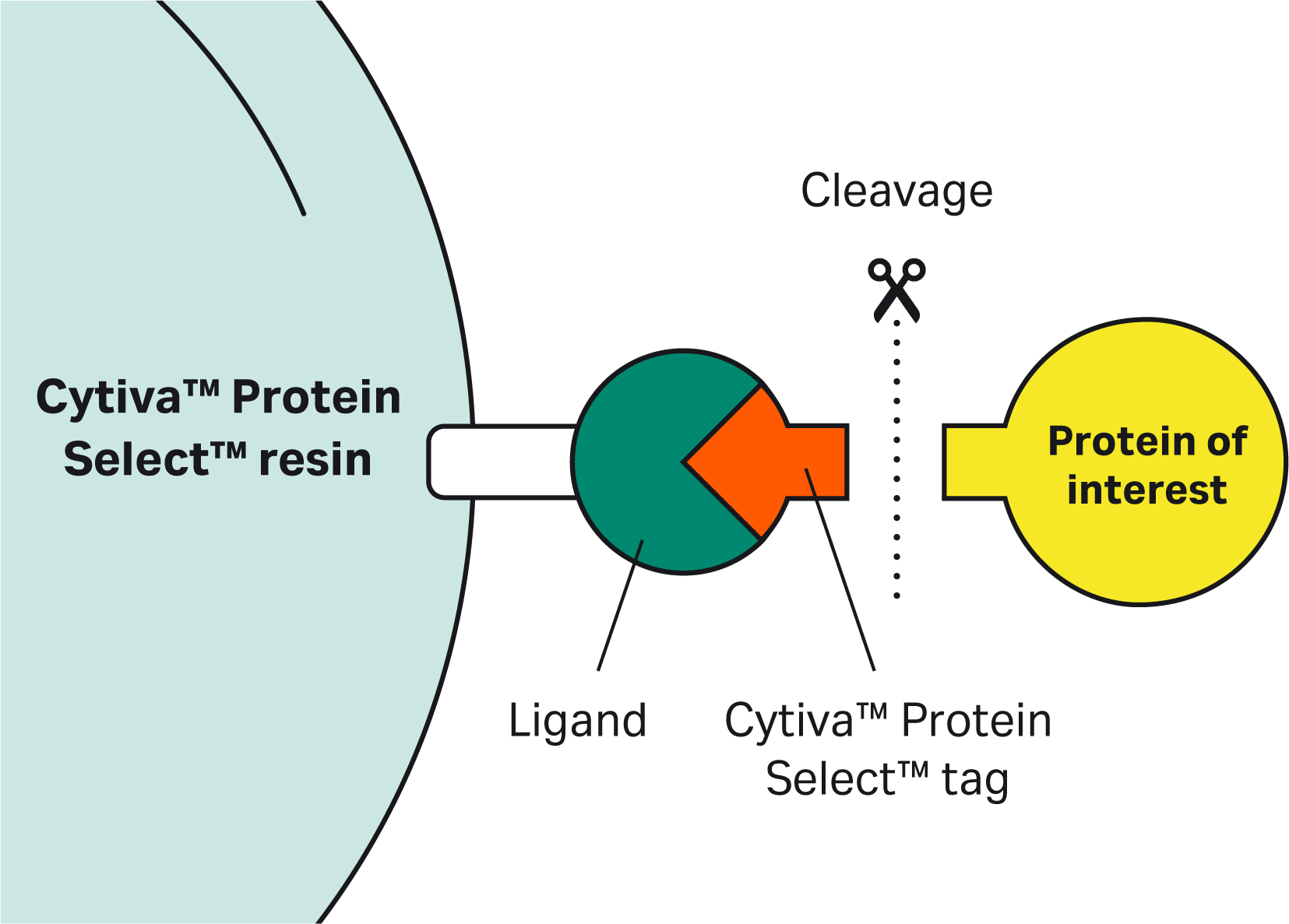
| 1 | What is the mechanism of self-cleavage? |
|
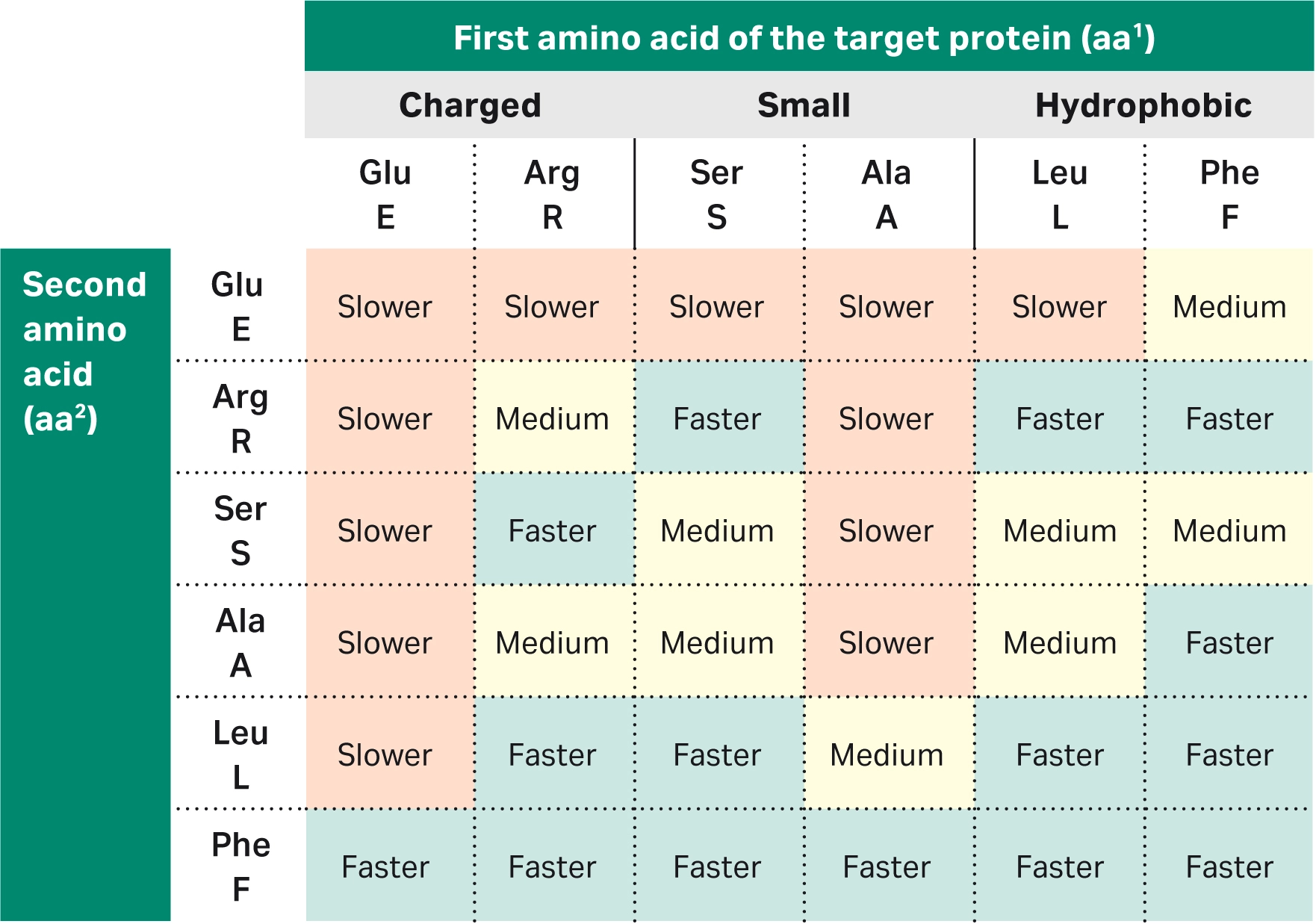
| 2 | How do the first three amino acids at the N-terminus affect the cleavage kinetics and cleavage time for proteins? |
The first three amino acids of the target protein heavily influence the cleavage kinetics, although the amino acid at the third position affects the cleavage rate to a lesser extent than the first two amino acids. Unfavorable combinations of the first three amino acids will result in slow tag cleavage and thus low target protein yield during purification.To see data on the impact of the two first amino acids on the cleavage efficiency, read Factors affecting cleavage efficiency when using Cytiva™ Protein Select™ technology. Briefly, we’ve found:
We suggest avoiding Pro in the first or second position of the target protein, as it causes an extremely slow cleavage reaction. |
| 3 | You mentioned the importance of amino acids in positions 1 and 2. What about the effect of the amino acid at position 3 at the N-terminus of the target protein? |
Position 2 has the largest impact on cleavage rate, followed by position 1. Position 3 also has an impact, but to a lesser extent — i.e., position 3 is not equally important to consider. That said, it is the combined properties of the three amino acids (positions 1–3) that defines the cleavage kinetics. Find more details in question 2 of this section. |
| 4 | What initiates the cleavage? | The cleavage occurs after the tagged protein binds to the resin. The reaction is spontaneous and does not require additional excipients or stimuli. |
| 5 | Does self-cleavage require reducing conditions or any special condition? | No, the cleavage reaction is spontaneous and does not require additional excipients or stimuli. |
| 6 | Does cleavage occur only after the sample loading and wash step? | No, cleavage will start as soon as the tagged protein is bound to the resin. The instructions recommend a maximum of 30 min sample loading and wash time, but one should do sample loading and wash as fast as possible to minimize loss of target protein. If you have a slow-cleaving protein, you might still have reasonable recovery with sample loading and wash lasting longer than 30 min. |
| 7 | How do I determine the length of the hold step? |
We recommend that you screen different hold times (using the same sample load (amount and volume) and same flow rate). You can do this using several columns in parallel or by running several hold and elution steps on one column. We recommend 4 h as a starting point for the hold step. If the target protein is slow cleaving, overnight incubation might be required. An increase of pH and/or temperature can increase the cleavage rate. |
| 8 | What cleaves the tag? Do we need to use special reagents or buffer while the tagged protein is bound on the resin? | No, you do not need special buffers or reagents for the purification process. The tag is self-cleaving, and the cleavage reaction is spontaneous and does not require additional excipients or stimuli. You can use one buffer for the whole purification process.
Suggested buffers:
We recommend using 20 to 50 mM buffer compound supplemented with up to 1 M salt, e.g., NaCl. |
| 9 | Why doesn’t the Cytiva™ Protein Select™ tag work on the C-terminus? | The tag binds to the ligand on the resin, then the tag-ligand complex folds and forms a self-cleaving complex. Cleavage occurs between the last amino acid of the tag and the first N-terminal amino acid of the target protein. This process cannot happen if the tag is at the C-terminus of the target protein. |
| 10 | Is there protease linked to the column? | No, there is no protease involved in the system. |
| 11 | How do post-translational modifications (PTMs) affect the cleavage yield? | PTMs can potentially affect the cleavage rate. In our experiments with purification of the glycosylated receptor-binding domain (RBD) of the SARS-CoV-2 spike protein, the protein cleaved as expected. See data file. |
| 12 | How does having a Met in the first position affect cleavage rate? Should the Met always be removed? |
No, not always. Met is naturally removed in about 50% of proteins during translation, and more often in eukaryotic systems than in bacterial cells. So even if it is the most abundant amino acid in a protein structure, it is most often removed. This processing is usually described in annotated protein databases. |
| 13 | What is the effect of having Gly in the first amino acid position? |
The examples we have seen indicate that Gly in the first position leads to slower kinetics, whereas Ser as first amino acid leads to medium to faster cleavage. See reference 2 for a comparison of the kinetics of a protein with Gly as initial amino acid with a faster-cleaving version. |
| 14 | Is there a correlation between binding affinity and cleavage kinetics? |
There is no direct correlation between binding affinity and cleavage kinetics. The binding affinity between the Cytiva™ Protein Select™ ligand and tag is at least in the low-nanomolar range and is independent of the target protein attached. |
Large-scale production and column packing
| Question | Answer | |
|---|---|---|
| 1 | Can the resin be used in GMP production? | Support and products for scaling up to clinical and commercial scale are available, including regulatory support files (cytiva.com/RSF). The resin is available in 1 L and 5 L units for packing in larger columns, such as HiScale™ or AxiChrom™ chromatography columns. Learn how to pack AxiChrom™ chromatography columns with Cytiva™ Protein Select™ resin in this article. |
| 2 | How do I scale up using a flow rate of 600 cm/h and sample load and wash time of 30 min? If your sample isn’t highly concentrated, it will take much longer than 30 min to load and wash the column. What column design would work best in this case? | We recommend using wide columns with a bed height of 10 cm. The Instructions for Use (1) recommends a flow rate of 600 cm/h during sample load and wash for up to 30 min. This is a strong recommendation for the first trial; however, for slow-cleaving target proteins, the flow rate during sample loading and washing can be decreased (e.g., to 400–500 cm/h), which will extend the time for sample load and washing. In such cases, increasing bed height is an option. |
| 3 | How do I pack Cytiva™ Protein Select™ resin in a column? |
Please refer to these instructions:
|
| 4 | Can I use Cytiva™ Protein Select™ resin for food applications such as enzyme purification? |
Cytiva™ Protein Select™ resin may be suitable for enzyme purification in food-related applications. A regulatory support file (RSF) is available to assist with regulatory assessments. However, please note that regulatory requirements can vary by region, and it is the user’s responsibility to ensure compliance with all applicable local regulations before use. |
Licensing
| Question | Answer | |
|---|---|---|
| 1 | Is there a licensing fee for using Cytiva™ Protein Select™ tag and resin? | No; upon purchase of the Cytiva™ Protein Select™ resin you receive a non-transferable limited license to make and have made biomolecules comprising the Cytiva™ Protein Select™ tag, solely for the purpose of purification of said biomolecules using the Cytiva™ Protein Select™ resin. Any other use will require a separate license from Cytiva. |
Ligand
| Question | Answer | |
|---|---|---|
| 1 | What is the nature of the ligand? Is it a small molecule, peptide, or something else? | Cytiva™ Protein Select™ technology uses a protein ligand produced in E. coli. |
| 2 | Do you have ligand samples available that I could use to develop and validate my own analytical method for detecting ligand leakage? |
Yes, samples are available. You’ll first need a restricted use license to get a sample. Contact your local sales representative, as they can help with both. |
Ligand leakage assay
| Question | Answer | |
|---|---|---|
| 1 | What is the ligand leaching detection method for Cytiva™ Protein Select™ resin? | A commercial ELISA kit for ligand leaching detection in Cytiva™ Protein Select™ resin is under development but not available at present. A restricted license for Cytiva™ Protein Select™ ligand is available for users who wish to generate antibodies for the assay developments. |
| 2 | What toxicity studies have you done for the ligand and tag? | Our toxicity assessments for Cytiva™ Protein Select™ resin and tag include an in silico sequence similarity study, an interaction study with blood components, and study of potential routes of elimination. Overall, these investigations indicate limited toxicity concern for process impurities from this technology. More information can be found in the regulatory support file (RSF) at cytiva.com/RSF. |
Multiple tags
| Question | Answer | |
|---|---|---|
| 1 | Does the system still work if I add tags upstream of the Cytiva™ Protein Select™ tag? If so, do you have recommendations for how to design the construct? |
You can add extra amino acids such as signal peptides or additional tags upstream of the Cytiva™ Protein Select™ tag to facilitate detection, purification, analysis, or solubility. This should not affect the system’s efficacy; you’ll still obtain native protein with no remaining tag amino acids after purification. You can find more information in the article Design flexibility of protein construct with Cytiva™ Protein Select™ tag, including how to design the construct with extra amino acids to:
|
| 2 | Will a higher-molecular-weight solubility tag (GST or MBP) in front of the Cytiva™ Protein Select™ tag influence the interaction between the Cytiva™ Protein Select™ tag and the ligand on the resin? That is, is it necessary to remove the added tag before binding to the ligand? | Although we have not tested this, it should be possible to have an MBP or GST tag upstream of the Cytiva™ Protein Select™ tag. It should still bind and cleave (note, however, that a potential issue with GST is its propensity to dimerize, which would affect the cleavage). You might also consider adding a linker (suitable for larger protein tags) between the two tags to reduce the risk of steric hindrance. |
| 3 | When adding a signal peptide or dual tag upstream of the Cytiva™ Protein Select™ tag, is there a limit to the size? Could you add a sequence of 90–100 aa, for example? And how would that affect binding or cleavage efficiency? | We are not aware of a size limitation of a dual tag or a signal peptide upstream of the Cytiva™ Protein Select™ tag. Things to consider are potential interactions of a dual tag, and a suitable linker between the tags to allow a certain degree of movement for the different parts of the fusion protein. Any effects of an additional tag on binding and cleavage could be tested against a single-tagged control protein. We have seen minor effects on cleavage rate (slightly slower), with an additional 40 aa affinity tag attached via a (GGSG)-linker. |
| 4 | Can a small tag, such as a His- or FLAG-tag, be cloned between the Cytiva™ Protein Select™ tag and the target protein? |
We have tested proteins with His-, Strep-tag II, and FLAG-tags upstream of the Cytiva™ Protein Select™ tag. They bind to the resin and cleave as expected. |
| 5 | Do you think the Cytiva™ Protein Select™ tag could help solubilize the protein? | The tag is rather small (about 4 kDa), so it will not help solubilization. You can place a solubilization tag upstream of the Cytiva™ Protein Select™ tag. You might also consider adding some linker amino acids between the tags. |
| 6 | Will adding a FLAG or histidine tag upstream of the Cytiva™ Protein Select™ tag prevent or affect capture by the ligand? | No. It is OK to have a FLAG or histidine tag upstream of the Cytiva™ Protein Select™ tag. We have tested proteins with other tags upstream of the Cytiva™ Protein Select™ tag and they worked well. |
Protein constructs
| Question | Answer | |
|---|---|---|
| 1 | Are there any sequences that can serve as positive or negative controls? | Yes. We have been using human IL-1β for positive and negative controls. The mature sequence contains a proline in the second position, which binds but cleaves poorly, so it can be used as negative control. The positive control has a proline-to-phenylalanine substitution, which cleaves relatively fast. |
| 2 | If the tag sequence is used in a mAb, does it matter if it is on the light chain or heavy chain? | This must be empirically tested. Note also that the kinetics of the cleavage reaction are likely to be different from that of monomeric proteins and will likely need some optimization. Generally, we recommend using dedicated affinity resins for mAb purification. (See also the question below on multimeric proteins.) |
| 3 | What is the impact of free cysteine? | Addition of cysteines to the media or the protein of interest should not impact binding and cleavage activity. |
| 4 | You observed that hydrophobic/aromatic amino acids in the second position may increase the rate of cleavage. Is this increase enough to cause a problem—for example, loss of target protein in the wash due to the speed of the reaction? | Low yield caused by fast cleavage is possible, especially if the sample application time is long. In that case, optimization of the purification method may be required to increase the yield. We recommend using a high flow rate during sample load/wash and/or concentrating the sample to reduce the overall time. |
| 5 | Do you have data on how this system works with multimeric proteins? | We don’t have much data on multimeric proteins. If the protein is a homo-multimer and all monomers carry a tag, purification may result in heterogeneous products due to multiple binding and cleavage events. In contrast, hetero-multimers with only one tagged monomer should yield more consistent results. So, purifying multimeric proteins with this resin and tag might result in low yield and/or heterogeneous protein. |
| 6 | Will the performance of the Cytiva™ Protein Select™ resin be hindered with a 60 kDa protein of interest? | No, a protein of 60 kDa will not hinder the performance of the resin. |
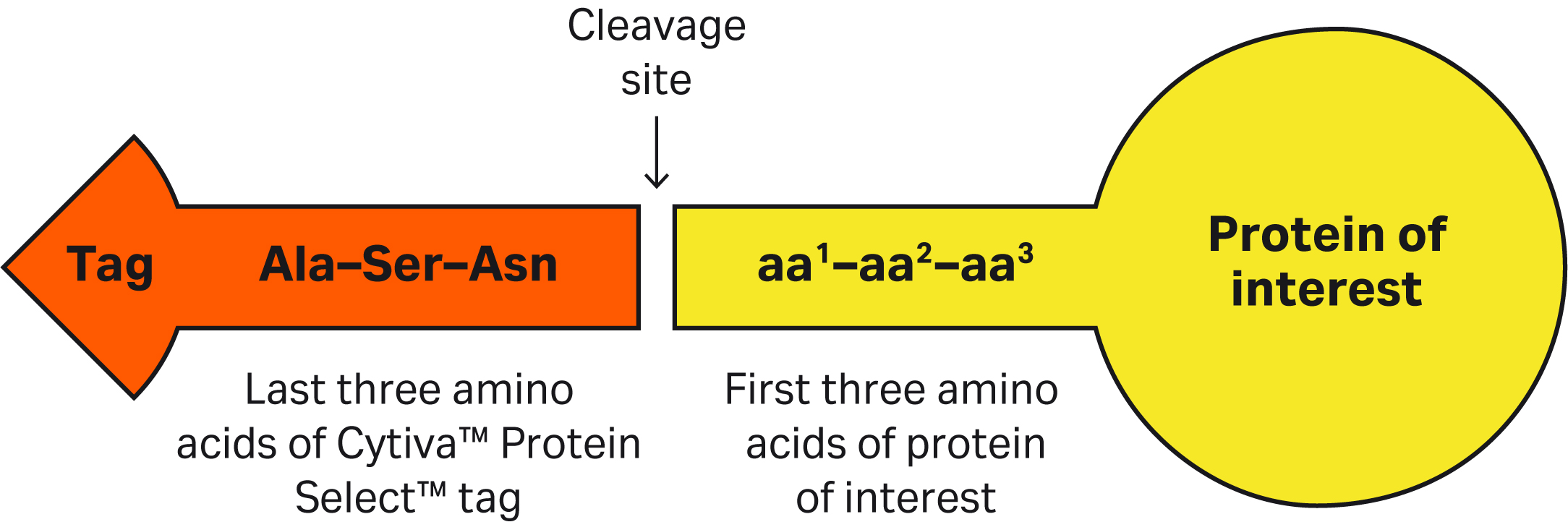
| 7 | If the first three amino acids of target protein contain sequences that might halt or slow the cleavage process and we don’t want to mutate the target protein, can we add a linker sequence (that includes all the recommended amino acids for efficient cleavage) between the C-terminus of the tag and the N-terminus of the protein? | Yes, one can insert amino acids between the C-terminus of the Cytiva™ Protein Select™ tag and N-terminal of protein. Cleavage will be after the last amino acid of the tag (Asn). Inserting Ala-Phe-Val or Phe-Arg-Val has shown fast cleavage for the proteins we have tested. Find more details in this article: Factors affecting cleavage efficiency when using Cytiva™ Protein Select™ technology. |
| 8 | The Cytiva™ Protein Select™ tag ends with Ala-Ser-Asn. What happens if we have the same sequence at or close to the N-terminus of our protein? Would we see unexpected cleavage? | Having an Ala-Ser-Asn sequence in your protein will not affect the cleavage position. After the tag binds to the ligand on the resin, the tag-ligand complex folds and forms a self-cleaving complex, wherein cleavage occurs after Asn of the Ala-Ser-Asn in the tag. |
| 9 | Is there any limitation regarding the use of further tags upstream of the Cytiva™ Protein Select™ tag? For example, how does size play a role (e.g., MBP tag)? Any recommendation here? Which tags have worked best so far? |
We have tested mainly epitope tags like polyhistidine, FLAG-tags, and Twin-Strep-tag. |
| 10 | If we have some small and nonpolar or negatively charged amino acids in the first, second, and third position of the protein, will the cleavage be total after 24 h? | No, some uncleaved protein will likely remain bound to the resin. If you want to improve the cleavage kinetics, you could also consider adding two additional amino acids (such as Ser-Leu). We have also tested for predicted protein domains and shifted the start of the domain (e.g., +1 or -1 from the intended start). In this way the sequence will still be "native". Find more details in this article: Factors affecting cleavage efficiency when using Cytiva™ Protein Select™ technology. |
| 11 | Does the size of the target protein affect the binding or cleavage efficiency? | The first three amino acids have a larger impact than the size of the protein. Very large proteins could potentially have steric hindrance during binding to the resin. |
| 12 | Does the system work with PEGylated proteins? | It should work with PEGylated proteins, but it's not something we have explored. |
| 13 | What are the protein sizes tested so far? | We have tested proteins between 15 and 100 kDa. |
| 14 | Your data file says the first three amino acids of the protein are critical for self-cleavage efficiency. Do you have examples of the most optimal amino acid sequences? | In our tests, proteins that have Ala-Phe-Val or Phe-Arg-Val as the first three amino acids showed fast cleavage rate. |
| 15 | Should we add linkers between tags like His-tag and the Cytiva™ Protein Select™ tag, and do you have guidance on that? |
Yes, preferably include a small, flexible linker, typically 4–8 amino acids like Gly-Ser (e.g., GGSG repeated once or twice). GGSG is what we recommend, but alternatives may also work. |
Purification conditions
| Question | Answer | |
|---|---|---|
| 1 | The data file says that 1 M is the highest NaCl concentration recommended for the buffer; what is the lowest NaCl concentration recommended? | Salt is not required for binding or cleavage, but a good starting point could be to add 150–300 mM NaCl to minimize unspecific interactions and increase purity. |
| 2 | Do you have experience using this affinity tag and resin with proteins at low expression levels — for example, 100 mg/L and a 200 L scale? We're wondering about loading time and the risk of losing protein of interest at flow-through. | Cytiva™ Protein Select™ needs a shorter sample load and washing phase because of the self-cleaving activity, so it’s best to avoid low-expressed proteins unless you can add a concentration step before binding.
For example, you could concentrate mammalian cell feeds by ultra-filtration to increase sample load. Concentration can also be done using other techniques than ultra-filtration, including ion-exchange chromatography. |
| 3 | You recommend 4 h or overnight for the hold step. Would leaving it for a longer period, such as one or two days, increase the cleavage? | For proteins with slow cleavage rate, a hold time of 1–2 d can increase cleavage and thereby improve yield if the target protein is stable during that time. |
| 4 | At 50°C or 60°C, is it possible to have efficient self-cleavage? | We don’t have data on cleavage at 50°C or 60°C. Theoretically, cleaving is faster at these temperatures, but the stability of the column hardware, resin, and the target protein are also important to consider. |
| 5 | Do you have any recommendations for batch purification? | We do not have a protocol for batch purification. Because tag cleavage occurs after the tagged protein is bound to the resin, we recommend fast sample application and wash. That is why we recommend purification using a packed column. |
| 6 | Is the sample loading and wash time limited to a maximum of 30 min? | The 30 min sample loading and wash time is a guideline to minimize risk of losses. However, the maximum loading and wash time depends on the cleavage kinetics of your target molecule. The impact of loading time is lower for a slow-cleaving protein. |
| 7 | How much of the sample should we expect to recover after purification? | The recovery depends on several factors, including the first three amino acids of the target protein (which affect cleavage rate), the sample loading and washing time, and the hold time. You can read an example here: Case study: process development for purification of recombinant proteins. |
| 8 | How can we improve the recovery? |
To improve recovery, first identify which purification step causes the loss. Fine tune the loading, washing, temperature, and buffer conditions to maximize the recovery. You can read more details in Optimizing protein recovery in the purification step using Cytiva™ Protein Select™ resin technology. |
| 9 | What sample preparation method should I use for a target protein expressed in E. coli? | Suspend expression E. coli pellet/cells in binding buffer (1 g cells per 5–10 mL buffer) prior to lysis (e.g., homogenization or sonication). Clarify the sample by centrifugation (we recommend starting with 50000 × g for 20 min). Immediately before applying it to the column, centrifuge and/or filter the sample through a 0.22 µm or a 0.45 µm filter. |
| 10 | The buffer recommended for Cytiva™ Protein Select™ resin is MES, Tris or PBS with pH 6–9, but how about adding low-concentration salt to the buffer to reduce nonspecific reactions, especially during sample application? | It is always good to have some salt to reduce nonspecific reactions. We recommend 20–50 mM buffer compound supplemented with up to 1 M salt (preferably NaCl), at a pH range of 6 to 9, which means one can use 0 to 1 M salt. |
| 11 | How efficiently does the tag bind to the ligand during sample application? At the maximum recommended sample application of ~600 cm/h, is it possible that some of the sample might end up in the flowthrough? | The affinity between the tag and the ligand is very high, and we see fast binding of tagged protein to the resin. A small amount of cleaved protein will be in the flowthrough and wash (this is also dependent on the first three amino acids of the protein, which affect the speed of cleavage). |
| 12 | If I purify protein from cell culture supernatant with a large sample volume and sample loading takes a long time, is there a risk that parts of the protein will be cleaved and elute before loading of the column is complete? | We recommend that sample application, including the wash step, is completed within 30 minutes. This is because if the target protein is fast cleaving, some loss of cleaved protein will occur during sample application. Consider sample concentration before purification. |
| 13 | Can we perform the purification at 4°C? | Yes, you can run the purification at 4°C (such as in a cold room) but cleavage will be slower, so you may need to increase the time for the hold step. |
| 14 | Why do the His-tagged and Cytiva™ Protein Select™ tagged proteins look like they’re different sizes after purification (on the SDS-PAGE gel shown in the data file)? | In the data file we showed a comparison of Cytiva™ Protein Select™ tag vs His-tag with TEV protease. The purified protein from the Cytiva™ Protein Select™ tag does not contain extra amino acids, while the one from His-tagged protein contains residual amino acids after TEV cleavage. We confirmed both molecular weights by LC-MS. See data file. |
| 15 | How do you prevent tag cleavage during the binding and wash steps? | The cleavage will start once the tagged protein is bound to the resin. This is why we recommend the sample loading and wash are done as fast as possible. |
| 16 | What is the composition of the elution buffer? Is it necessary to exchange elution buffer for dialysis or size exclusion chromatography, for example? | The protein will elute in the buffer that was used for wash after sample loading. There is not a dedicated elution buffer. |
| 17 | Can wash buffers be applied to remove unwanted impurities? | Yes, after sample loading you can use different buffer(s) for washing out unwanted impurities. |
| 18 | Can we use protease inhibitors in buffers? | Yes. Pefabloc and EDTA worked well in our purification. |
| 19 | What is the pH dependency of the cleavage reaction? | The cleavage is rather insensitive to pH at range 6–-9. For some proteins, switching from, say, MES pH 6.2 buffer for sample loading to Tris pH 8.5 for washing buffer might increase the cleavage rate, but this is of course protein dependent. Find more details in this article: Factors affecting cleavage efficiency when using Cytiva™ Protein Select™ technology. |
| 20 | Can you comment about nonspecific binding on the resin? | We have not seen any particular issue with nonspecific binding, but it is always a good idea to include some salt in the buffer to reduce nonspecific binding. |
| 21 | Are the concentrations listed in the “Resin stability and compatibility” section of the Instructions for Use (1) for operation or for storage? And are they upper limits, or could a higher concentration of these buffers be used? | The “Resin stability and compatibility” section of the Instructions for Use (1) lists the buffers and additives that Cytiva™ Protein Select™ resin can be stored in for 1 week. Some of these are not suitable for operation. During purification we suggest these buffers:
|
| 22 | Does cleaving rate impact protein purity? | No, cleaving rate does not impact purity of eluted protein. |
| 23 | How do you prove traceless cleavage? | The eluted protein has no traces of tag amino acids, confirmed by mass spectrometry analysis. |
Purified proteins
| Question | Answer | |
|---|---|---|
| 1 | In the data file you showed mass spec data on the scaffold protein. Have you characterized the analysis artifact peak? | Yes, it is an acetonitrile adduct (+42) and likely a neutral loss of H20 (-18). |
| 2 | Does the protein stay active once purified with Cytiva™ Protein Select™ technology? | Yes. Protein activity is retained after purification with Cytiva™ Protein Select™ technology. Data published in reference 2 shows that SARS-CoV-2 spike protein RBD has a high degree of binding to RBD monoclonal antibodies (measured by biolayer light interferometry) when purified by immobilized metal affinity chromatography (IMAC) or Cytiva™ Protein Select™ resin. They also showed results on an artificial bispecific T-cell engager (BiTE) designed to link T cells with cells expressing epithelial cell adhesion molecule. When purified with IMAC or Cytiva™ Protein Select™ resin, the BiTE showed similar CD4+ and CD8+ T-cell activation profiles, as well as cytotoxic activities (characterized using a viability assay monitoring real-time changes in cellular impedance) (2). |
Resin
| Question | Answer | |
|---|---|---|
| 1 | What is the base matrix of the resin and why was this base matrix chosen? | The resin is based on an established high-flow agarose matrix with a particle size of ~ 60 μm. This is one of Cytiva’s modern affinity purification resins. We chose it based on its resolution and scale up capabilities. |
| 2 | What are the formats available for Cytiva™ Protein Select™ resin? |
Cytiva™ Protein Select™ resin is available in 25 mL, 100 mL, and 500 mL packs of bulk resin and in HiTrap™ 1 mL and 5 mL columns (HiTrap™ Protein Select™ columns), which are well suited for research and process development work. Instructions are supplied for packing Cytiva™ Protein Select™ resin in Tricorn™ and HiScale columns. |
Tag
| Question | Answer | |
|---|---|---|
| 1 | How big is the Cytiva™ Protein Select™ tag? | The Cytiva™ Protein Select™ tag is roughly 4 kDa in size. It is made of 36 amino acids. For more information about this tag, visit cytiva.com/protein-select, choose your application, and fill in the form. The sequence of the tag will be sent by email to you immediately. |
| 2 | How strong is the affinity of the Cytiva™ Protein Select™ tag to the resin? | The affinity between the Cytiva™ Protein Select™ tag to the ligand on the resin is very high. Measurements of surface plasmon resonance (SPR) signals on a Biacore™ instrument show very high affinity and an extremely low dissociation rate. |
| 3 | How can I obtain the sequence of Cytiva Protein Select tag? | Visit cytiva.com/protein-select, choose your application, and fill in the form. The sequence of the tag will be sent by email to you immediately. |
Upstream
| Question | Answer | |
|---|---|---|
| 1 | How do I Integrate Cytiva™ Protein Select™ tag into an expression vector? |
To generate a recombinant protein with the Cytiva™ Protein Select™ resin it is essential to design a DNA construct (a plasmid) that positions the Cytiva™ Protein Select™ tag at the N-terminus of the protein. Refer to the article Integrating Cytiva™ Protein Select™ tag into an expression vector for guidelines and recommendations for the design and generation of such constructs. |
| 2 | Have there been instances of the tag cleaving in the cell culture broth (via protease or cofactor)? |
Tag truncation can occur during expression or purification, regardless of the tag system used. Some truncations can impair the target protein's ability to bind to the resin. You can try the following to minimize tag truncation:
|
| 3 | If the tag is self-cleavable, what is the stability of the tag inside prokaryotic and eukaryotic cells? | The self-cleavage can only occur after binding to the resin. Of course there is the risk that the tag itself can be cleaved by other proteases in the sample, e.g., if the sample is stored for an extended time at room temperature. |
| 4 | Can you recommend cell culture conditions? | No, as this will be very dependent on the protein of interest and the expression system. |
| 5 | How do users determine titer of their cell culture when using this technology? | That depends on the protein of interest (POI). We suggest that the titer determination is done on the POI or on additional tags, if used. |
| 6 | What vector type do you suggest for the plasmid? | Any vector should be fine—or at least, we haven’t found any compatibility challenges to date. See question 1 in this section for more details. |
| 7 | Are yeast or insect cell expression systems suitable for the Cytiva™ Protein Select™ system? | We haven’t tested the technology with yeast or insect cell expression systems, but we have no reason to think it wouldn’t be successful. |
| 8 | What titer and yield should we expect from the upstream process? Is there a difference in performance between His-tag and Cytiva™ Protein Select™ tag in protein expression? | The answer depends on the expression system and target protein. We have tested expression of native proteins and of His-, FLAG-, and Strep-tagged proteins in comparison to the Cytiva™ Protein Select™ tag. Those studies showed similar expression levels with and without addition of the Cytiva™ Protein Select™ tag. |
| 9 | Does the size of the Cytiva™ Protein Select™ tag relative to that of the target protein affect expression or activity? | In the proteins we’ve tested so far, we haven’t seen any major effect of the tag on target protein expression levels in bacterial or mammalian systems, nor on the functionality after purification and tag removal (2). |
| 10 | Which signal peptides did you use successfully? | We have successfully used Ig-kappa, CD36, and IL-2 signal peptide sequences. |
| 11 | Are antibodies to the Cytiva™ Protein Select™ tag available? | No antibodies to the Cytiva™ Protein Select™ tag are available now. They may be available in the future. In the meantime it is possible to add, e.g., a FLAG or His-tag upstream of the Cytiva™ Protein Select™ tag for detection. |
| 12 | Can we insert the tag into any expression plasmid? | Yes, you can add the tag sequence to your protein of interest and in your vector of choice. This can be done in many different ways, such as tag and gene synthesis, cloning of genes or inserts by PCR or restriction digestion, or Gibson assembly. You can also buy synthetic genes from external vendors. See an example in this article: Integrating Cytiva™ Protein Select™ tag into an expression vector. |
| 13 | Can you produce tagged protein in cell culture medium containing serum? | Yes. We have tried a few different media formulations, and we prefer serum free because it's less complex. But the system works with cell media that contain serum as well. |
| 14 | Would this work in cell-free systems? | We haven't evaluated that, but in theory it should work in a cell-free system. |
| 15 | Do you provide the DNA sequence for the Cytiva™ Protein Select™ tag? | We provide the amino acid sequence of the tag, but the optimal DNA sequence will depend on the expression system and should be optimized together with the protein of interest. There are several codon optimization tools available to improve the rate of translation by overcoming limitations associated with host cell codon usage and the abundance of transfer RNA (tRNA). Although we have not seen an impact on the tag’s performance based on DNA sequence, codon optimization has the potential to cause changes in the target protein’s conformation and functionality.
Many external vendors can produce plasmids optimized for yield based on amino acid sequence and expression system. |
| 16 | Can any signal peptide be used in combination with the Cytiva™ Protein Select™ tag? Other than removing the initial Met of the tag, are there signal peptide constraints or limitations? |
In principle, any signal peptide can be used with the Cytiva™ Protein Select™ tag. We have tested just a few. Removal of the tag’s initial Met residue is not required. |
| 17 | Does the cleavage start within the expression system? |
No, it does not. Cleavage starts only after your target protein is bound to the resin (after which a self-cleaving complex is formed). Cleavage will not occur during expression or sample preparation. |
References
- Cytiva™ Protein Select™ resin: Affinity Chromatography Resin Instructions for Use. Cytiva, 29711002 AA: V19; 2023:6
- Clifford R, Lindman S, Zhu J, et al. Production of native recombinant proteins using a novel split intein affinity technology. J Chromatogr A. 2024;1724:464908. doi:10.1016/j.chroma.2024.464908