By Jon Lundqvist, Cytiva and David Haylock, VivaZome.
We demonstrate a scalable workflow for the isolation of exosomes—a type of extracellular vesicle with significant potential for targeted drug/gene delivery. For research and development and diagnostic purposes, ultracentrifugation is an established technology to separate and purify exosomes. However, ultracentrifugation is not easily scalable. We describe an evaluation of purification workflows from three different cell culture supernatants. The workflows we describe are suitable for scale-up and combine tangential flow filtration (TFF) for the concentration of exosomes followed by gentle size exclusion chromatography (SEC) with our next-generation Cytiva™ superSEC resin. We found that:
- Cytiva™ superSEC resin enriched exosomes from impurities compared to legacy Sepharose™ CL-2B resin—a well-established resin for exosome purification—after the initial TFF step.
- High productivity, scalability, and single-use compatibility were observed for the superSEC resin.
- Scaling up the SEC resin volumes using the superSEC resin was clearly shown with shorter purification cycle times than used in existing SEC protocols for the purification of exosomes.
Introduction
Exosomes are a subset of extracellular vesicles (EVs) that play important roles in intercellular communication and various physiological processes. Enrichment and concentration of exosomes from biological samples are essential for studying their functions and clinical applications and for drug delivery and therapeutic production. However, the enrichment and concentration of exosomes from cell culture supernatants (CCS) containing a mixture of nanosized EVs is a major challenge for exosome manufacture. Furthermore, although ultracentrifugation and density gradient separation are beneficial techniques for the concentration of exosomes for research purposes, they are not easily scalable.
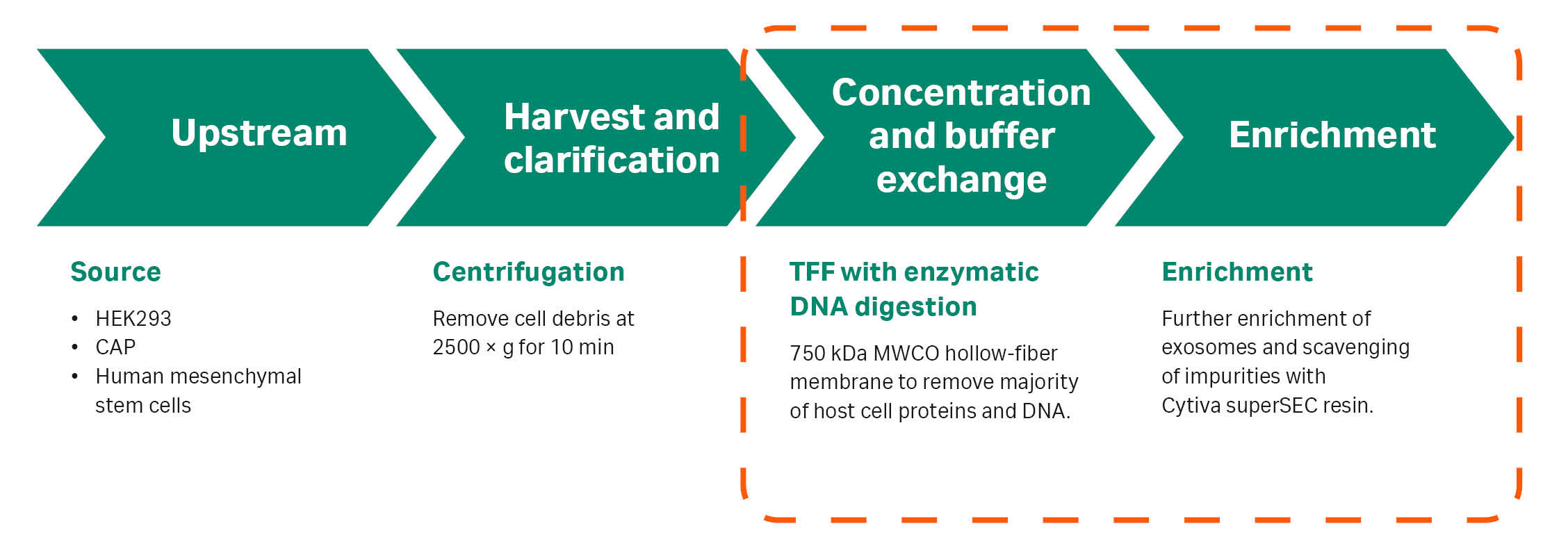
Tangential flow filtration (TFF) and size-exclusion chromatography (SEC) are two commonly used methods for the enrichment of exosomes from complex biological fluids. Combining these well-known techniques, which are gentle methods for exosome purification, allows the removal of most of the host-cell protein (HCP) and DNA (Fig 1). In addition, such a workflow gives high recoveries and minimizes the loss of biological activity of the purified exosomes.
Exosomes from different cell types cultured in different media and conditions are known to display different glycosylation patterns and surface proteins—this affects not only biological activity but also how exosomes interact with filtration membranes and chromatography resins. We developed a gentle and scalable workflow for three cell lines and evaluated the results in three different cell culture supernatants (CCS):
- Human mesenchymal stem cells (hMSC)
- Cytiva’s amniocyte production cells (CAP™)
- Human embryonic kidney cells (HEK293)
The workflow tested is described in Figure 1. In the SEC phase of the workflow, we compared the performance of two SEC resins—Cytiva™ legacy Sepharose™ CL-2B and the next-generation Cytiva™ superSEC resin.
Fig 1. The TFF/SEC workflow for exosome purification evaluated in this study (orange dashed line).
Sepharose™ CL-2B SEC resin is established for the enrichment of exosomes from smaller impurities (1). In labs purifying exosomes at small-scale, low-flow chromatography columns or gravity columns packed with Sepharose™ CL-2B are often used to remove protein and DNA impurities. The fractionation range of next-generation Cytiva™ superSEC resin is comparable to Sepharose™ CL-2B, but the resin can also be packed in large-scale columns. In addition, it has flow properties that allow you to quickly run a full SEC chromatography cycle, including regeneration. This gives a downstream workflow that is scalable for processing volumes of exosomes needed for therapeutic purposes. The processing steps are compatible with GMP guidelines and with single-use equipment.
Capto™ Core multimodal (mixed mode) chromatography resins offer an alternative for gentle size-based scavenging of impurities from large entities like EVs, allowing the purification of large sample volumes. Regeneration, of these resins resin requires 30% isopropanol for regeneration unlike superSEC resin, which offers the advantage of regeneration with water in isocratic mode.
Production of exosomes
Three cell lines expressing exosomes were used in the three examples presented:
- RoosterBio human mesenchymal stem cells (hMSC), RoosterNourish Expansion Media, and RoosterCollect EV collection media were used to produce exosome-rich cell culture supernatant, used at VivaZome.
- Cytiva’s amniocyte production cell line (CAP™) producing GFP-exosomes provided by Evox.
- Human embryonic kidney 293 (HEK293) producing exosomes, internally produced by Cytiva.
Harvest and clarification
All cell supernatants were centrifuged to remove cell debris (10 min at 2500 × g).
Initial exosome enrichment and concentration (TFF including degradation of host DNA with benzonase).
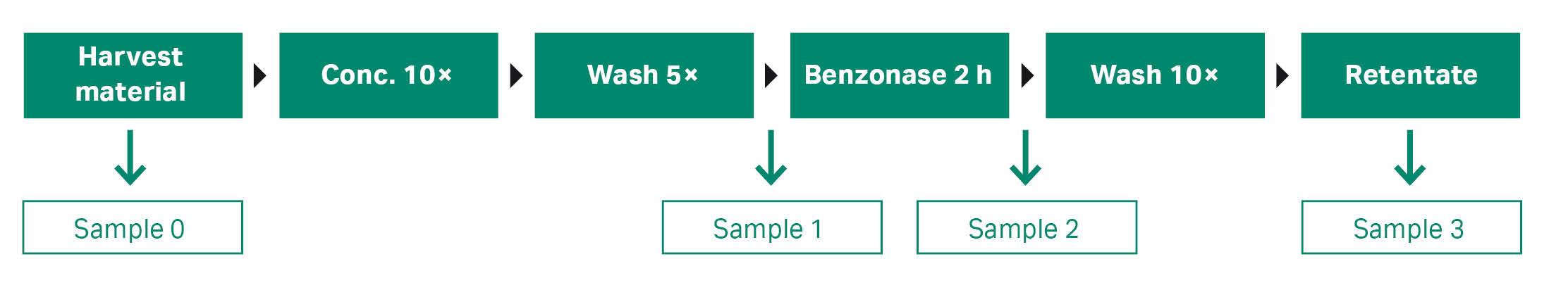
In all three examples, exosome concentration and buffer exchange were carried out using an ultrafiltration (UF) cartridge with MWCO 750 kDa (Cytiva) on an ÄKTA flux S TFF system at 4000 shear rate (TMP 0.4 bar, 5.8 psi, 0.04 MPa). A 10-fold concentration and 5-fold diafiltration were performed using PBS, pH 7.4. At the end of the process, benzonase (2 kU +1 mM MgCl2) was added to the concentrated exosome retentate and incubated for 2 h. Finally, a diafiltration (10-fold with PBS) was performed to remove any degraded DNA residues, see Figure 2. The exosome retentate was concentrated 10-fold and then collected for further purification using columns packed or prepacked with superSEC resin.
Fig 2. TFF workflow for concentration, buffer exchange, and benzonase treatment of exosome solution.
Size exclusion chromatography (SEC)
Cytiva™ superSEC and Sepharose™ CL-2B resins were used for SEC. In example 1, both resins were packed in Tricorn™ 10/200 according to the standard packing instructions. The superSEC resin was also packed in a HiScale™ 26/40 column with a bed height of 20 cm according to standard packing instructions. In examples 2 and 3, two HiScreen™ superSEC columns were coupled in series to give 20 cm bed height in a small-scale purification. HiScale™ 26/40 column was packed with superSEC resin for larger-scale purification. Sample volumes and flow velocities are described in Table 1. All runs were performed on the ÄKTA pure™ 25 chromatography system.
Table 1. Running conditions used on columns packed with superSEC resin; ÄKTA pure™ 25 chromatography system was used for the runs
| Example* | |||||
|---|---|---|---|---|---|
| 1 | 1 | 2 and 3 | 1 | 2 and 3 | |
| Resin | Sepharose™ CL-2B |
superSEC |
superSEC | superSEC |
|
| Column | Tricorn™ 10/200 |
2 × HiScreen™ (7.7/100 bed size) | HiScale™ 26/40 |
||
| Bed height (cm) |
20 | 20 |
20 | 20 |
20 |
| Bed volume (mL) | 17 | 17 | 9.4 | 106 | 106 |
| Buffer | PBS, pH 7.4 | ||||
| Sample volume | 2 | 2 | 1 | 12 | 15 |
| Sample as percentage of bed volume | 12.5 | 12.5 | 10.6 | 11 | 14 |
| Flow rate (mL/min) | 0.15 | 0.5 | 1 | 3 | 4 |
| Flow velocity (cm/h) | 11 | 38 | 129 | 34 | 45 |
| Chromatography cycle time (min) | 150 | 50 | 20 | 50 | 45 |
Analytical methods
Spectrophotometric analysis (UNICORN™ software) at 260, 280, and 490 nm absorbance online on ÄKTA pure™ 25 chromatography system.
Nanoparticle tracking analysis (NTA). The samples collected from the concentration and enrichment steps were analyzed in NTA using a Z-NTA ZetaView (Particle-Metrix) instrument to determine particle concentration and size distribution.
ELISA assay analysis: The Human CD63 ELISA Kit (EH95RB, Thermo Fisher Scientific) was used according to the manufacturer’s instructions.
Total protein concentration: Pierce™ BCA Protein Assay Kit (23225, Thermo Fisher Scientific)
DNA content: The Qubit ssDNA Assay Kit (Q10212, Thermo Fisher Scientific) was used to measure the ssDNA content in the samples.
Western blotting (Wb): The expression of canonical exosome proteins such as tetraspanins (CD9, CD63, CD81) and luminal proteins such as ALIX, TSG101, and syntenin-1 were quantitated by Wb analysis in example 1.
Antibodies for transmembrane protein CD81 and the cytosolic protein TSG101 were used in example 2.
Exosome morphology: Transmission electron microscopy (TEM) was performed on a concentrated exosome preparation to assess vesicle integrity and aggregation using a JEM-2100 (Jeol) microscope following a well-established protocol for negative staining in Example 1. Scanning electron microscopy (SEM) was employed to examine the morphology and size of the exosomes, in addition to any presence of impurities in examples 2 and 3.
Packing of Sepharose™ CL-2B and superSEC resins
Sepharose™ CL-2B resin has flow properties that only allow for packing under gravity flow. Instructions are found here:
Cytiva SuperSEC resin is suitable for packing in chromatography columns.
For research-scale columns follow these instructions
For large-scale column packing follow these instructions
Cytiva™ superSEC resin is also available prepacked in ReadyToProcess™ columns via our Custom ReadyToProcess offering or prepacked in columns for research use. Contact your local Cytiva representatives for further information and guidance
RoosterNourish expansion media, and RoosterCollect EV collection media were used to produce exosome-rich cell culture supernatant. The work was performed by VivaZome.
hMSCs were used to produce exosome-rich cell culture supernatant (CCS). Multiple exosome manufacturing companies and academic research groups are working with hMSCs as a source of exosomes. hMSCs are the most common producer cell type used for clinical trials.
A TFF hollow-fiber membrane with molecular weight cutoff (MWCO) of 750 kDa (73 cm2 surface area) cartridge (Cytiva, UFP-750-E-H42LA) was used in the diafiltration step. The procedure is described in Materials and methods. Digested DNA species and small nucleic acids were removed. Nucleic acids (host DNA content) were analyzed by the Qubit ssDNA analysis kit (Thermo Fisher Scientific) on the pre- and post-benzonase media/permeate from the ÄKTA flux™ S TFF system. The media pre-benzonase treatment showed a nucleic acid concentration of 24 ng/mL whereas the sample that was to be used for the SEC runs after treatment with benzonase had undetectable levels of nucleic acids (the lower limit on the DNA analysis is 1 ng/mL). A 10-fold concentration of exosomes with 100% recovery (no loss of target species) was obtained.
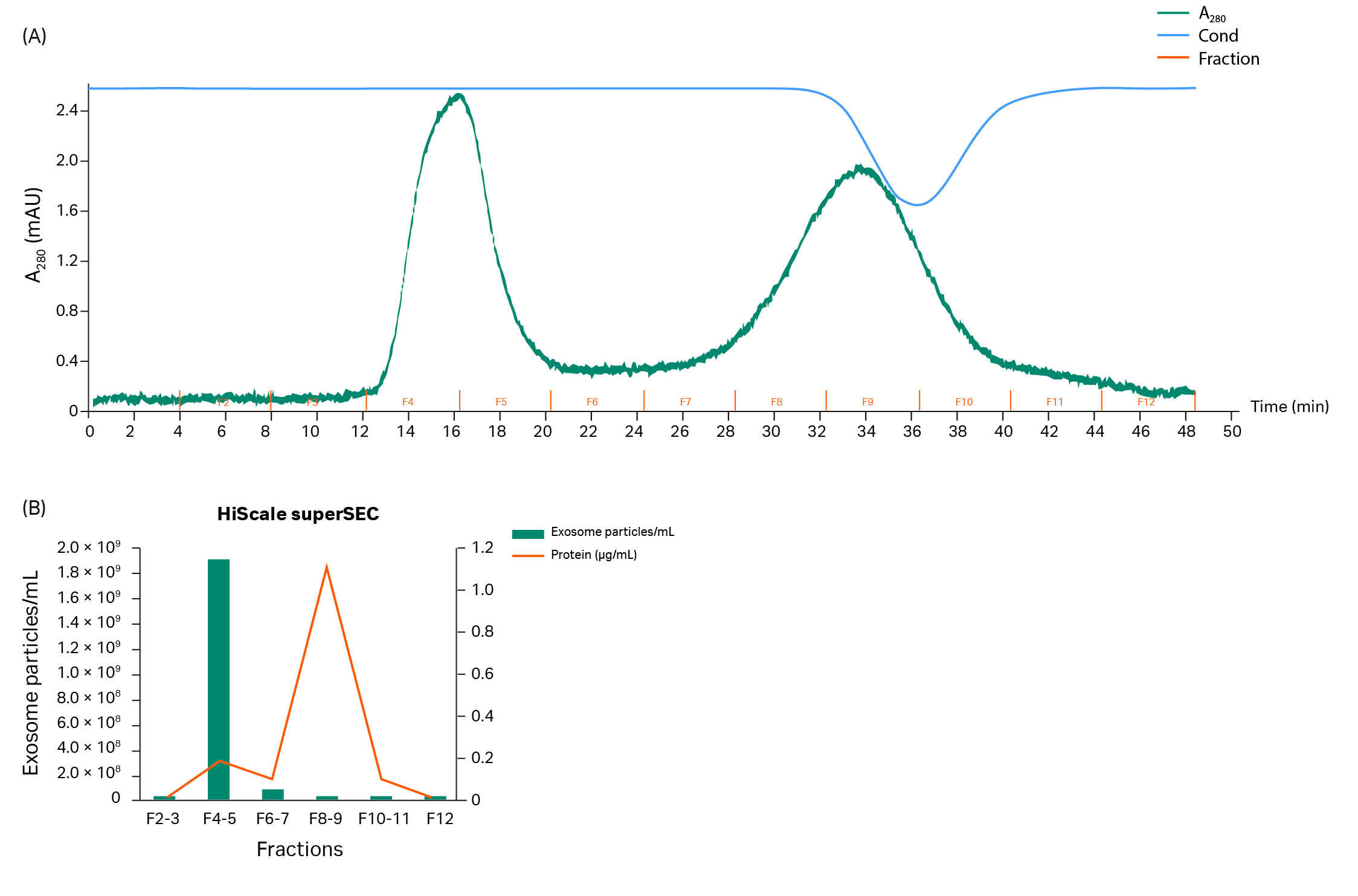
In the SEC step, Sepharose CL-2B (commonly used today) and the next-generation superSEC resins were compared in a Tricorn™ 10/200 column with a bed height of 20 cm (for running conditions, see Table 1 in Materials and methods). Different sample volumes (0.5 to 5 mL) were tested (data not shown). We found that a 2 mL sample volume (12.5%) was the most relevant choice for optimal performance and the chromatograms from both resins are shown in Figure 3 A–B. Both resins give a resolution that removes impurities that were not initially removed by TFF.
The hMSC-exosome concentration in the sample material after TFF was 2.2 × 1010 /mL (10-fold concentrated from the starting material). Fractions were collected from the SEC runs and analyzed with NTA, see Figure 3 C–D. Fractions of 4 mL volume each were collected, peak 1 (F3; early peak) and peak 2 (F5; the later peak) seen in Figure 3. The 4 mL from fraction peak 1 collected from the superSEC resin run contained 5.17 × 109 particles/mL, or 2.07 × 1010 in total, representing 47% of the starting number of particles. Figure 3 C–D shows that hMSC-exosomes are typically most concentrated in one of the collected fractions with the Sepharose™ CL-2B and superSEC resins. Notably, in both cases, in those fractions where hMSC-exosomes are most concentrated, there is a relatively low level of protein contamination (analysis with BCA assay).
Fig 3. Representative chromatograms from purification runs of hMSC with (A) Tricorn™ 10/200 packed with Sepharose™ CL-2B and (B) superSEC resins, respectively. F = fractions of 4 mL. Left peaks contained hMSC-exosomes while protein and DNA impurities were found in the right-hand peaks. Analysis shows particle and protein analysis of fractions collected from runs with Tricorn™ 10/200 packed with Sepharose™ CL-2B (C) and superSEC resins (D), respectively.
Based on the data and the benefits with higher flow/pressure properties for superSEC compared to Sepharose™ CL-2B resin, the superSEC resin is an improvement on Sepharose CL-2B. The vesicle recovery is similar but the superSEC resin has higher productivity if factors such as speed, bead rigidity, and scalability are considered. With the conditions used in this study, the run time on the superSEC resin was only 50 min with a flow velocity of 38 cm/h as compared to the much longer 180 min run time with Sepharose™ CL-2B resin using a velocity of 11 cm/h, (Fig 3 A–B). A shorter run time is a significant advantage for processing larger volumes of starting material.
Scale-up of superSEC resin packed in a HiScale™ 26/40 column
For the larger scale experiment with the HiScale™ 26/40 superSEC column (20 cm bed height), we performed four consecutive runs with a 12 mL sample load of the TFF-concentrated material. The maximum hMSC-exosome recovery with the superSEC resin was 66%, with a concentration of 4.56 × 1010 exosome particles/mL collected (Fig 4). The run time was 50 min with a flow velocity of 34 cm/h.
Fig 4. (A). Chromatogram from superSEC resin packed in HiScale™ 26/40 column run. (B). Particle concentration and protein concentration in each fraction.
Exosome structure and morphology and Western blotting
Fractions containing the highest number of hMSC-exosomes from runs with the HiScale™ column were pooled and assayed for exosomal content and morphology while exosomal proteins were determined by Wb.
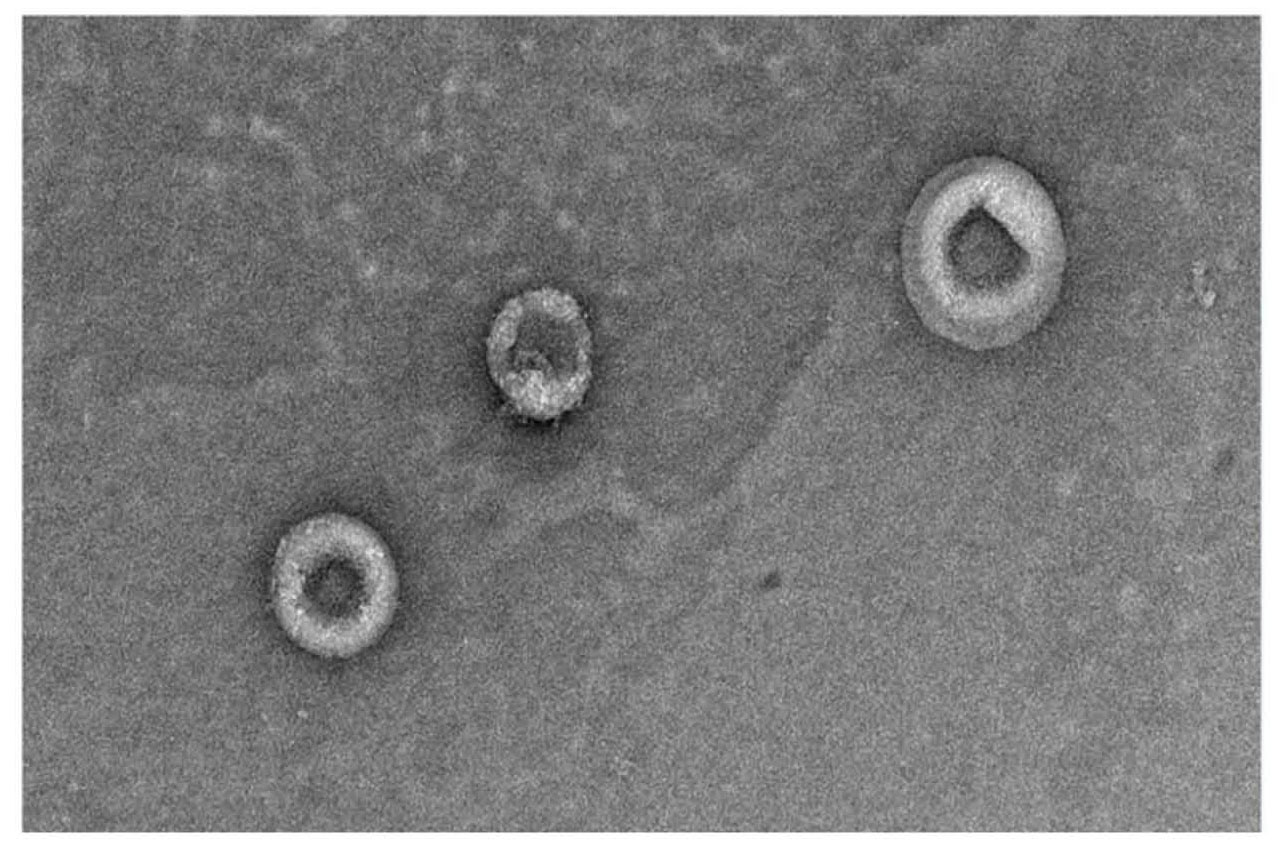
Transmission electron microscopy (TEM) was performed to assess the structure and integrity of exosomes purified with the column. A representative image of intact exosomes with their characteristic classical cup-shaped morphology structure and morphology is shown in Figure 5.
Fig 5. TEM image showing intact hMSC-exosomes fixed with glutaraldehyde and stained with uranyl acetate post-purification with superSEC resin (magnification 20 000×).
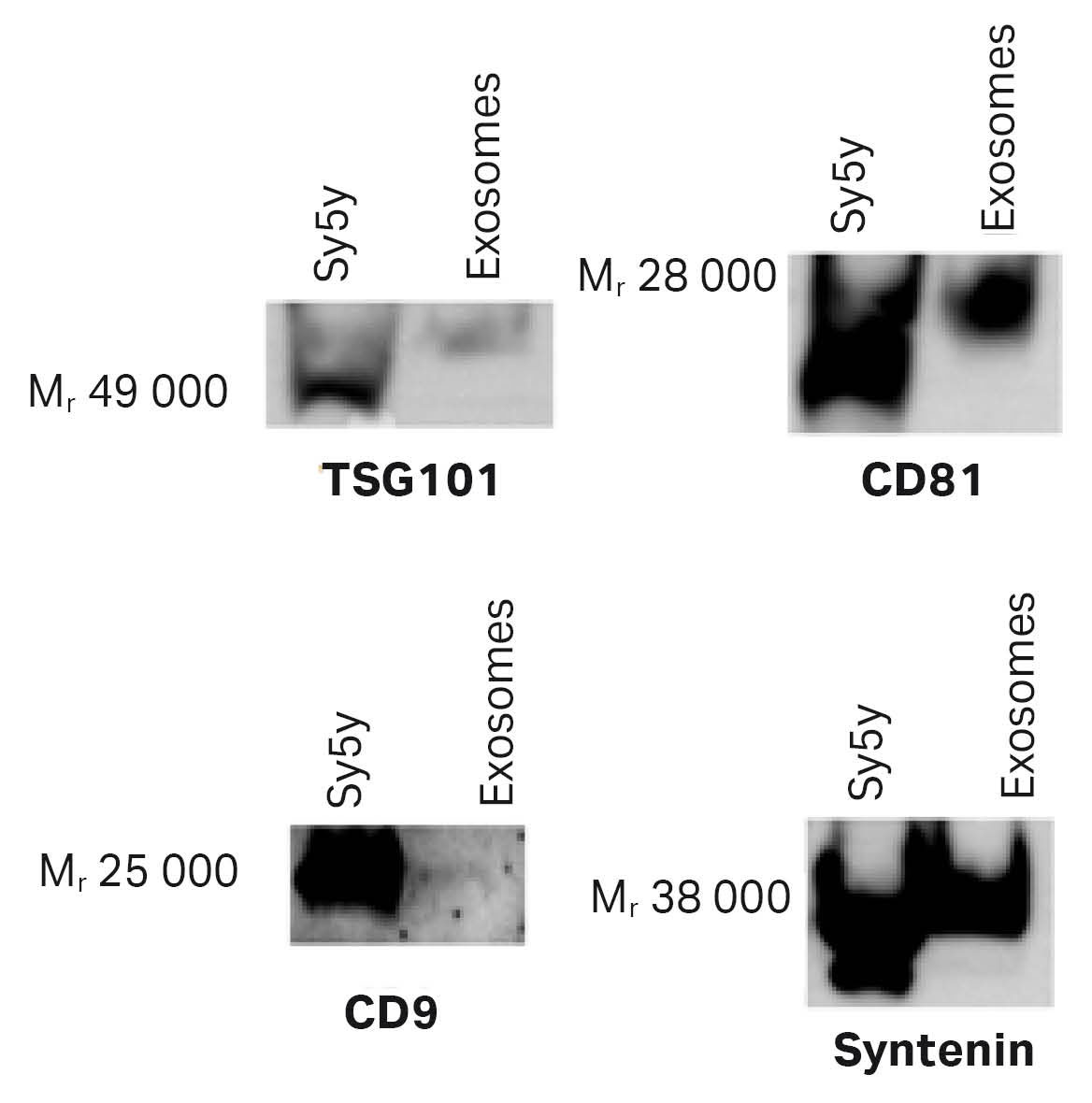
Images from the Wb analysis (Fig 6) confirmed that hMSC- exosomes expressed CD9 (albeit at low levels), CD81, syntenin, and TSG101. Expression of each of these target proteins on hMSC-exosomes is less than that observed in the positive control Sy5Y, where twice as much protein was loaded per lane.
Fig 6. Representative Western blots showing the presence of the exosome-specific proteins in the exosomes purified using a HiScale™ column packed with superSEC resin. The amount of protein loaded was 1.5 µg of exosomes and 3 µg of Sy5y exosome positive control.
In this workflow, the Cytiva's amniocyte production (CAP™) cell line was used to produce exosomes with a green fluorescent protein (GFP) tag linked to CD63 on the surface of the exosomes for fluorescence detection. To establish the reproducibility of the TFF-SEC protocol, two separate enrichments of GFP-exosome CAP™ CCS were performed with TFF followed by purification on the superSEC resin. As in example 1, two scales of purification were performed—two HiScreen™ columns, and a HiScale™ 26/40 column with 20 cm bed height, see Table 1 in Materials and methods. Recovery rates and exosome concentrations including particles (NTA), CD63 (ELISA), total protein, and ssDNA content analysis were measured. Wb and SEM were performed as a final confirmation of the identification of particles in the fractions collected.
A sample load of 250 mL of CAP™ CCS was applied to a TFF hollow-fiber membrane (MWCO 750 kDa) and filtration was performed according to the method described in Materials and methods. The sample (25 mL) was collected after the TFF step. Approximately 97% of the DNA and 92% of the protein were removed. According to analysis with NTA, the yield of particles retained after TFF was 56% of the starting particle concentration. With CD63 ELISA analysis, the exosome recovery was 43%.
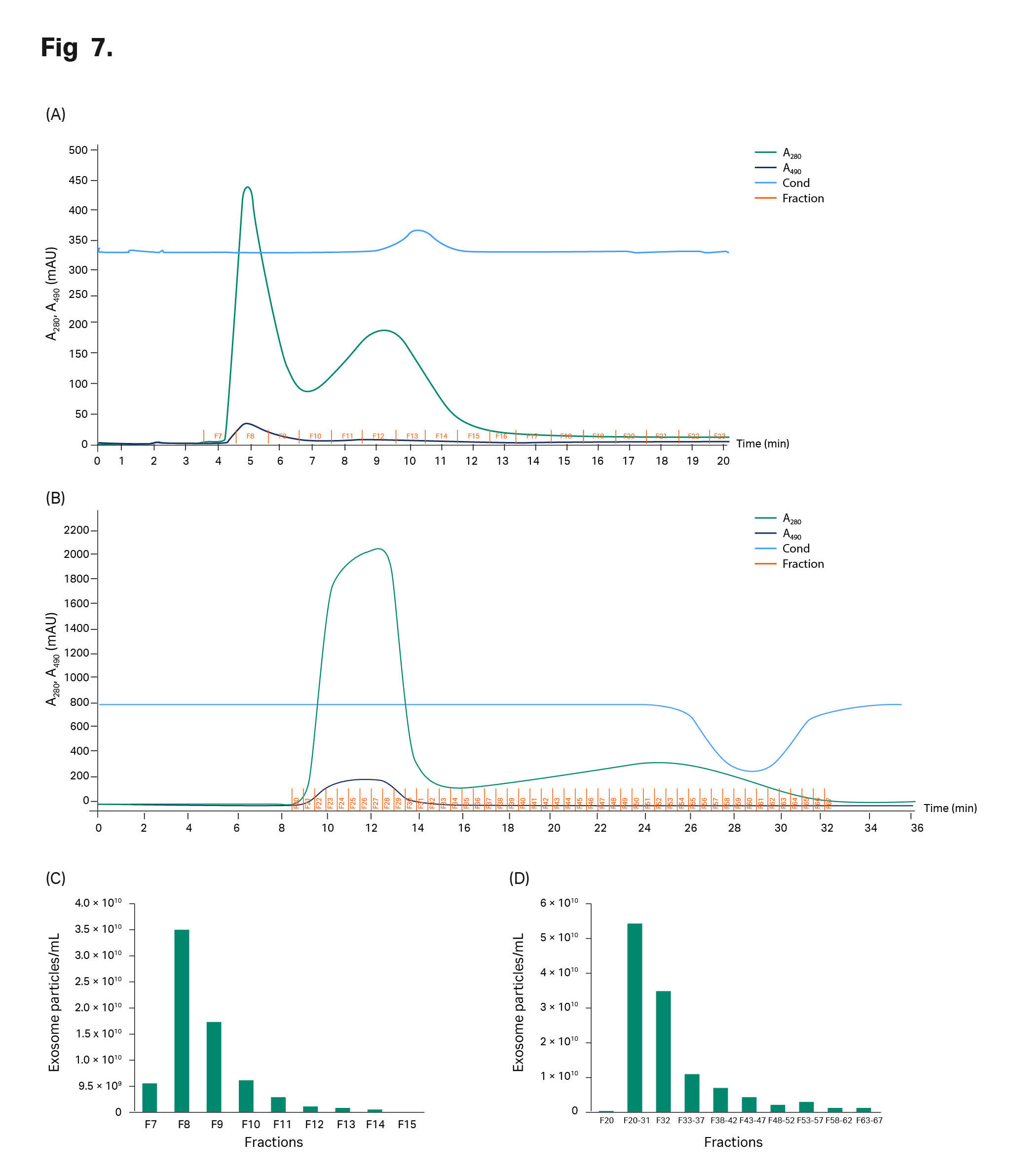
TFF retentate was then applied to the columns—1 mL was applied to the HiScreen™ superSEC column and 15 mL was applied to the HiScale™ column. The chromatograms are shown in Figure 7. The A490 nm curve showed that GFP-exosomes were eluted in the first peak from both column formats and that no A490 nm detection was seen in the second peak. The run times on the HiScreen™ and HiScale™ superSEC columns were 20 min (flow velocity 129 cm/h) and 45 min (flow velocity 45 cm/h), respectively (Fig 7). With equilibration and cleaning of the columns, the process time for one run would be 1 and 2 h, respectively.
Fractions from the chromatography runs were collected and analyzed with NTA, ELISA CD63, BCA, and Qubit ssDNA assay kit. With particle measurement (NTA, see Fig 7) of the fractions collected from SEC chromatography runs, the exosomes are retained in the first peak as confirmed by A490 nm detection in the chromatograms in Figure 7 A–B and ELISA CD63 (data not shown). Protein content analysis with BCA assay on peak 2 shows that more protein is removed with superSEC resin (Figure 7 C–D), which indicates that TFF cannot be used by itself if all host protein is to be removed. The results are also in line with the results obtained with the purification of exosomes from hMSC in example 1.
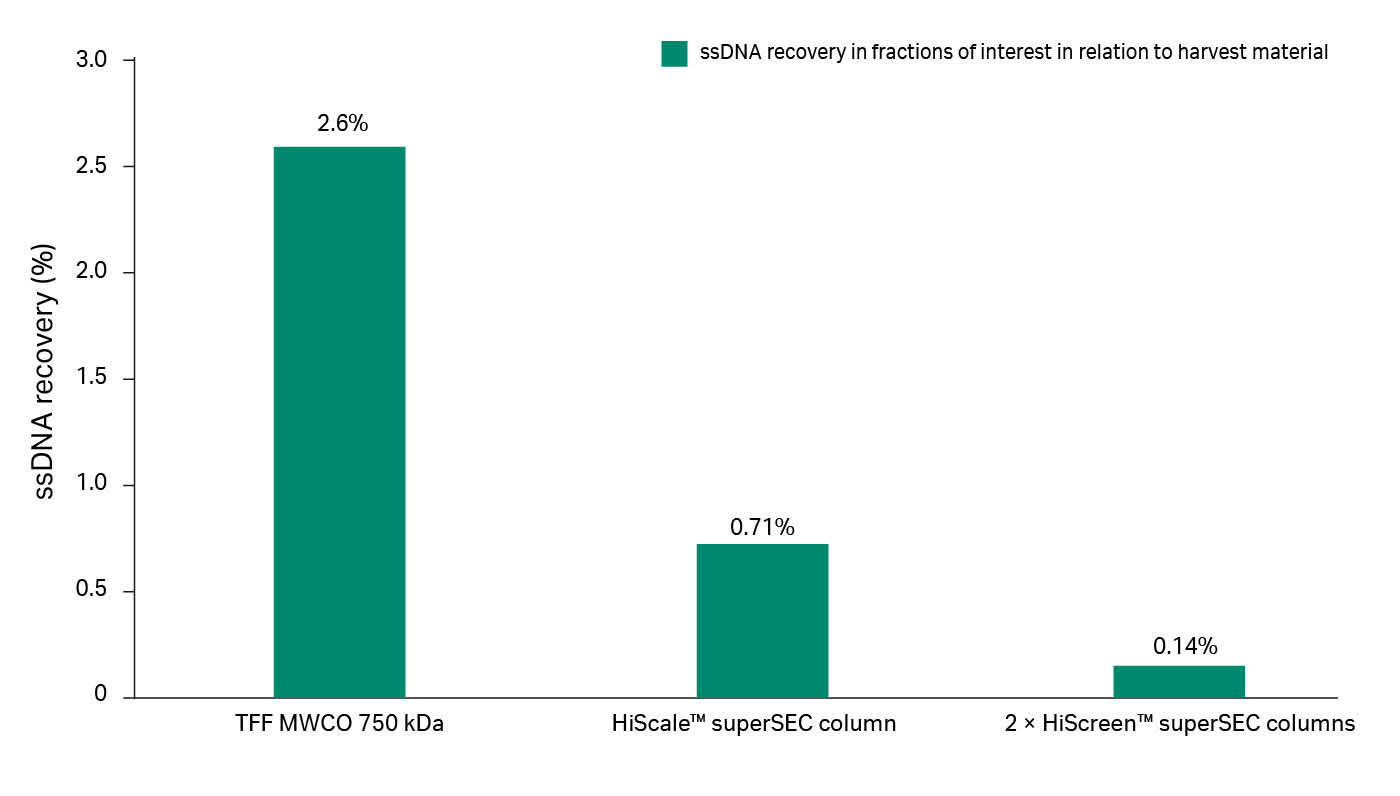
The results from the HiScale™ column run confirmed that exosome and protein contaminants can be separated in a larger scale as well. The benzonase treatment during TFF is effective and improves the removal of host DNA (Fig 8)—97% of DNA residues from the exosome sample were removed after the TFF step using benzonase treatment whereas only 40% was removed without benzonase. The superSEC resin purification step improves the DNA removal from exosome fractions even further, which confirms that superSEC is a valid alternative as a second step in exosome purification. The analysis with NTA also shows that the mean size (150 to 160 nm) of the particles did not differ through the process.
To further confirm that the exosomes are present in the collected fractions, Wb analysis with antibodies for CD81 and TSG101 and electron microscopy analysis using SEM was performed. Wb results (Fig 9) confirmed that these vesicles expressed CD81 and TSG101 even though TSG101 only was detected in the TFF retentate probably due to low concentrations in SEC fractions.
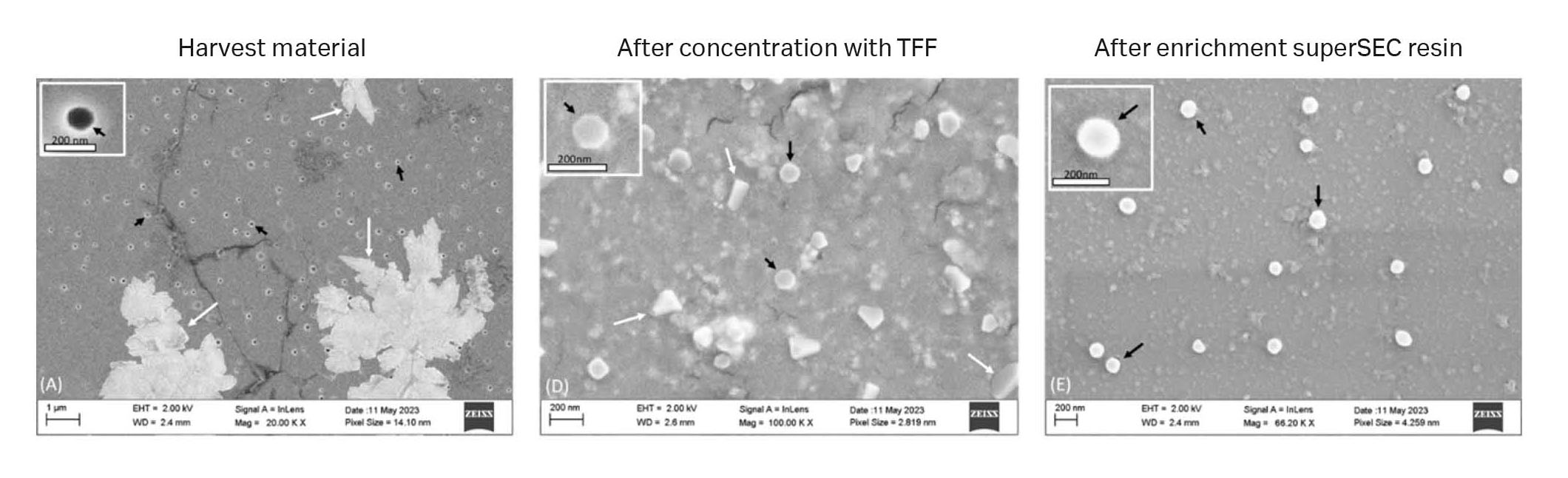
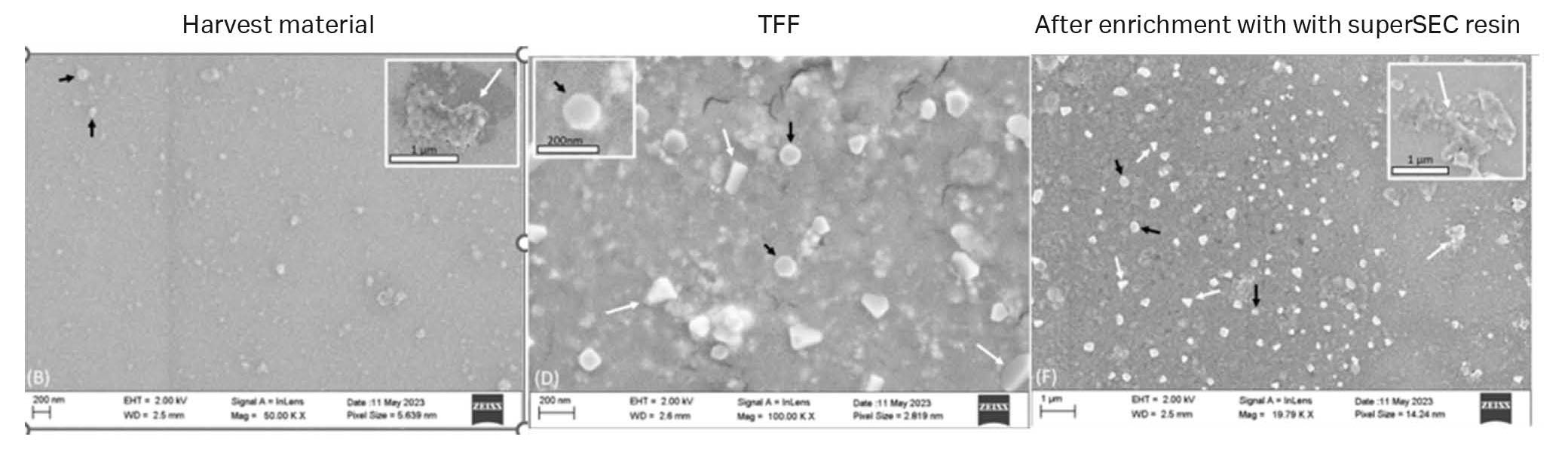
Images from SEM analysis (Fig 10) showed the presence of vesicles with classical cup-shaped morphology and that the TFF-SEC workflow removed impurities.
Fig 7. Chromatograms from the CAP™ CCS run on (A) HiScreen™ superSEC columns in series and (B) superSEC resin packed in HiScale™ column. Sample: TFF retentated GFP exosomes. (C) Particle concentration and protein concentration in each fraction from HiScreen™ and (D) HiScale™ superSEC columns.
Fig 8. The results of the ssDNA assay shows the removal of DNA after concentration and purification of GFP exosomes from CAP™ supernatant.
Fig 9. The Wb results for CD81 indicated the presence of CD81 in all steps of the isolation workflow, except for GFP exosomes from CAP™ cells after the HiScreen™ superSEC column run.
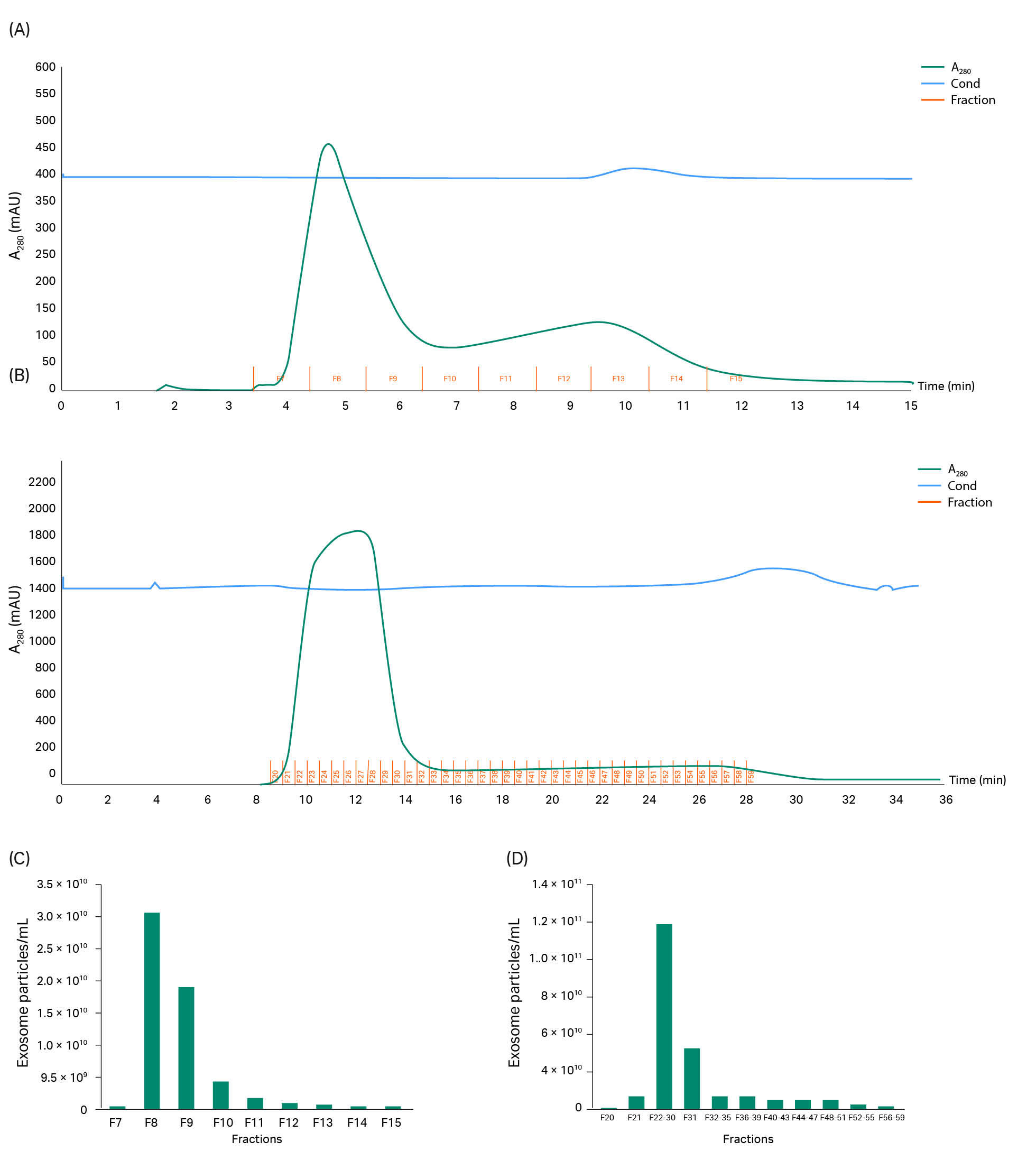
Fig 10. SEM images from different steps of the first isolation series of GFP-exosomes from CAP™ cells. Black arrows indicate exosome-like structures and white arrows indicate non-exosome structures. (A) Harvest material, (B) TFF 750 kDa MWCO retentate. (C) After HiScale™ superSEC column purification.
In this example, we purified exosomes from a HEK293 cell line that was internally produced at Cytiva. A HEK293 CCS sample of 250 mL was first loaded to the TFF hollow-fiber membrane (with 750 kDa MWCO) to remove HCP and DNA (according to the method described in Materials and methods). As with the two other examples of CCS, a benzonase treatment was applied, but no DNA analysis was performed on the HEK293 samples. Such as for the hMSC and CAP™ sample purifications, we deployed two scales of purification with superSEC resin (see Table 1 in Materials and methods). Recovery rates and exosome concentration including nanoparticle tracking analysis (NTA), CD63 (ELISA), and total protein analysis were measured. Wb and scanning electron microscopy (SEM) were performed as a final confirmation to detect the exosomes.
Approximately 92% of the protein was removed (BCA assay analysis). According to analysis with NTA, the yield of particles retained after TFF was 74% of the starting particle concentration. With the CD63 ELISA assay, the exosome recovery was 54%.
TFF exosome sample was then applied to the two column scales—1 mL of sample was applied to the HiScreen™ superSEC column and 15 mL was applied to the HiScale™ column. In the chromatograms in Figure 11 A–B, two peaks can be seen, as when loading hMSC-exosome and CAP™ TFF samples. The run time on the HiScreen™ and HiScale™ columns were also the same as for CAP™ TFF samples since the same flow velocity was used, 20 min (flow velocity 129 cm/h) and 45 min (flow velocity 45 cm/h), respectively, (see Fig 11). With equilibration and cleaning of the columns, the process time for one run would be 1 and 2 h, respectively.
Fractions from the chromatography runs were collected and analyzed with NTA, ELISA CD63, and BCA assay. In Figure 11 C, it is clear, based on NTA data that the exosomes are retained in the first peak, which was also confirmed by ELISA CD63 analysis (data not shown). The recovery of exosome particles after HiScreen™ and HiScale™ column runs were 51% and 61%, respectively. Also in this example, protein is removed using superSEC resin (Fig 11 C–D). The analysis with NTA also shows that the exosome particle means size (150 to 160 nm) did not differ throughout the process.
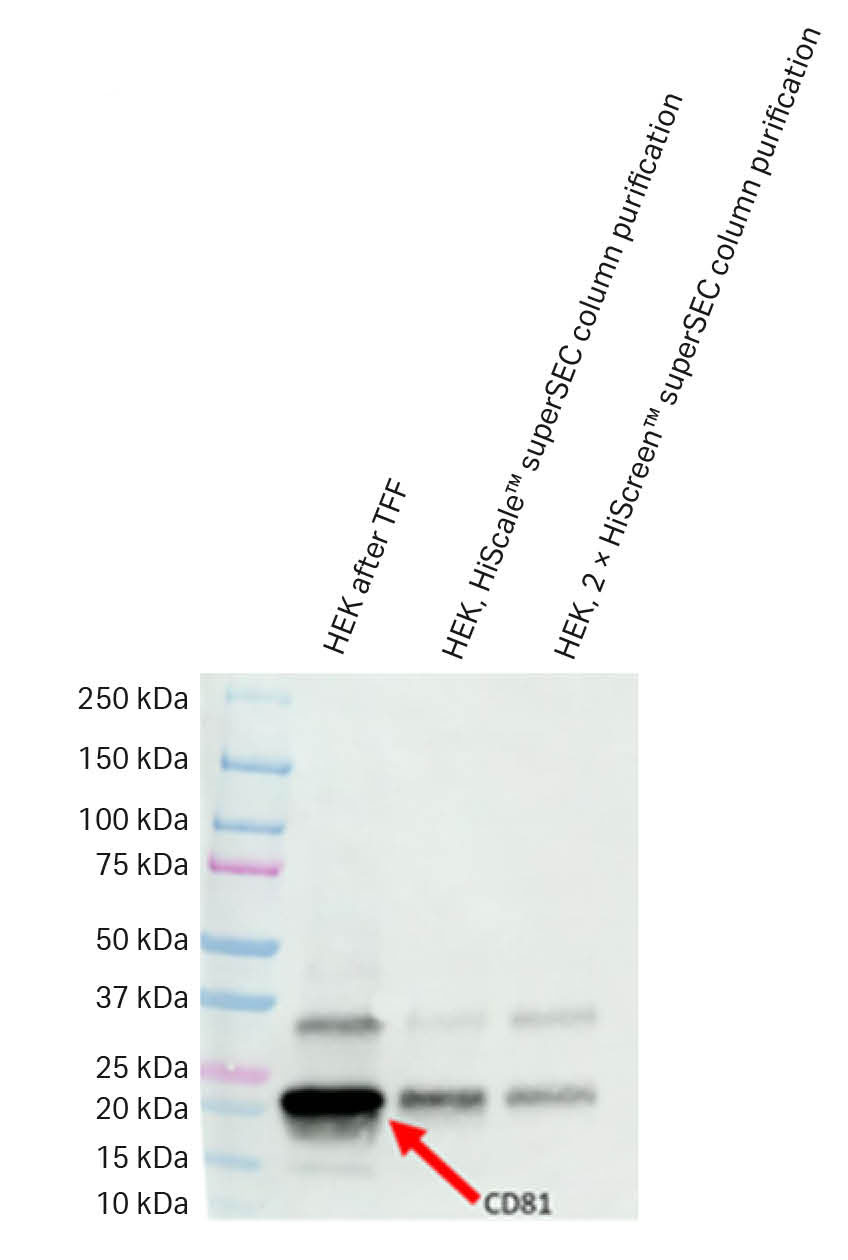
We performed Wb analysis (Fig 12) with antibodies raised against CD81 and TSG101. SEM analysis (Fig 13) was performed on fractions from the superSec runs in HiScreen™ and HiScale™ columns to confirm exosomal content.
The SEM images showed the presence of vesicles with classical cup-shaped morphology and that the different steps removed impurities. The Wb analysis confirmed that these vesicles expressed CD81 and TSG101 even though TSG11 only was detected in the TFF retentate due to low concentrations in fractions after SEC runs.
Fig 11. Chromatograms for (A), HEK293 CCS purified with HiScreen™ superSEC columns and (B) in a HiScale™ column packed with superSEC resin. Particle concentration and protein concentration in each fraction from (C) HiScreen™ and (D) HiScale™ column runs.
Fig 12. The Wb results for CD81 indicated a presence of CD81 in all steps of the isolation workflow.
Fig 13. SEM images of the purification of exosomes from HEK293 cell culture supernatant.
The results from the three CCS examples showed that Cytiva™ superSEC resin is well-suited as an exosome purification step after TFF. The advantages, besides the removal of smaller contaminations such as proteins, are improved pressure-flow properties, which allow for easy packing of the resin in large-scale columns and rapid SEC cycle time. This decreases the process cycle time by hours compared to using, for example, Sepharose™ CL 2B resin. It also provides you with the opportunity to perform several SEC cycles within a reasonable time frame.
Cytiva™ superSEC resin is a complement to the well-established multimodal chromatography method provided by Capto™ Core resins for exosome purification. Although Capto™ Core resins allow for larger sample volumes than SEC, the advantage with SEC is the opportunity it gives to collect any fraction from the chromatographic purification and the opportunity the elute all impurities, regenerating the resin for another chromatographic cycle using an isocratic gradient.
In addition to process intensification, the downstream exosome workflow with TFF and SEC, using Cytiva™ superSEC resin, shows an improved purity index than SEC with Sepharose™ CL-2B resin. The purity index is the ratio between the number of exosome particles compared to the amount of protein and DNA impurities. The performance regarding the enrichment of exosomes and removal of impurities has been shown with the three samples from three different CCS. We conclude that the purity index increases using Cytiva™ superSEC resin for two different scales and the three CCS.
We found that the separation of exosomes from protein during the SEC step was good. Vesicles pass through the resin early and protein is held up and collected in late fractions. The combination of exosome concentration with TFF and the flow properties of the superSEC resin lets you use a relatively small chromatography column, by performing repeated chromatography cycles.
In conclusion, purification with TFF followed by SEC is a gentle method to scavenge HCP and DNA from exosomes. The Cytiva superSEC resin allows for quick chromatography cycle time and the opportunity to perform several SEC chromatography cycles, within a limited time frame. TFF for initial removal of most impurities and sample concentration allows for effective further impurity reduction employing SEC.
- We have not tested many flow rates, but we have observed that 4 mL/min on a HiScale™ 26/40 column with 20 cm bed height (106 mL) gives > 50% exosome recovery. The amount of sample we loaded at 12.5% of CV gave the best resolution. Initially, we tried to 25% of CV sample loading but resolution was affected by the higher sample load.
- We have run the HiScale™ 26/40 column with 20 cm bed height with a flow rate of 4 mL/min (45 cm/h) using a load of 15 mL (14.2%) with good results. We have not tested higher flow rates with the HiScale™ column due to lack of sample, but you can probably go up in flow rate based on our data from runs at the smaller scale (data not shown). We ran a flow rate of 1 mL/min (128 cm/h) on two HiScreen™ columns coupled in series (20 cm bed height, 9.4 mL bed volume) with good results as well.
- Zhang X, Borg EGF, Liaci AM, Vos HR, Stoorvogel W. A novel three step protocol to isolate extracellular vesicles from plasma or cell culture medium with both high yield and purity. J Extracell Vesicles. 2020 Jul 14;9(1):1791450. doi: 10.1080/20013078.2020.1791450.
The data in this application note has been generated in collaboration with VivaZome Therapeutics Pty Ltd in Australia and Evox Therapeutics Ltd in the UK.
The following have contributed to this application note:
Cytiva: Jon Lundqvist, Inger Salomonsson, Peter Guterstam, Jagan Billankanti, Sofie Hellsten, and Vincent Goijenfalk.
Vivazome: David Haylock and Mitch Shambrook.
CY38458-150224-AN