This article demonstrates the ability of PuReTaq™ Ready-To-Go™ PCR Beads to deliver low background amplification along with polymerase chain reaction (PCR) efficiency, flexibility, and reproducibility in real-time PCR.
Ready-To-Go™ PCR Beads are ambient-temperature stable, complete single-dose reactions. The beads offer flexibility while yielding robust and reproducible PCRs optimized for use with endpoint polymerase chain reaction (PCR) and with a number of real-time PCR systems including intercalating dye, TaqMan™, and MGB™ Eclipse Probe System.
Cytiva developed Ready-To-Go™ PCR Beads to minimize nonspecific amplification by eliminating nucleic acid contamination. Nonspecific amplification generates spurious bands and background smears and generally reduces the overall efficiency of PCR. Ready-To-Go™ PCR Beads eliminate background amplification by employing PuReTaq™ DNA polymerase and other high-purity reagents. The resulting amplification of target amplicons is more robust and reliable than with other systems. Formulated with low MgCl2 concentration, the beads also provide flexible reaction control, so you can customize both performance and background fluorescence in your reactions. In addition, bead manufacture includes rigorous real-time PCR quality control test methods to ensure that each lot meets specifications for reproducibility and provides only the lowest possible levels of contaminating nucleic acids.
This article demonstrates the ability of Ready-To-Go™ PCR Beads to deliver low background amplification with PCR efficiency, flexibility, and reproducibility. It also presents results from side-by-side comparisons of Ready-To-Go™ Beads with other master mix formulations in identical real-time PCR assays. In these tests, the beads yielded more robust growth curves with lower threshold cycles, indicating more efficient PCR.
Products used
| Ready-To-Go™ PCR Beads | |
| 0.2 mL tubes, plate of 96 | 27955701 |
| 0.2 mL tubes, 5 plates of 96 | 27955702 |
| 0.5 mL tubes, 100 reactions | 27955801 |
| 0.2 mL tubes, 96 reactions | 27955901 |
Other materials required
- Lambda DNA PCR Control Kit (Cytiva, discontinued)
- TaqMan™ custom design primers/probe system (Applied Biosystems)
- MGB™ Eclipse Probe System (Epoch Biosciences)
- Smart Cycler™ (Cepheid)
- Roto-Gene 3000™ (Corbett Research)
- ABI PRISM™ 7700 and 7900 HT Sequence Detection System (Applied Biosystems)
- Sterile, biology-grade water
- MgCl2
- GelStar™ nucleic acid gel stain
- AmpliTaq™ Gold DNA Polymerase (Applied Biosystems)
- Platinum™ Pfx DNA Polymerase (Invitrogen Life Sciences)
Protocol
PCR setup
- In the pre-PCR room, remove the required number of tubes (each tube contains one Ready-To-Go™ PCR Bead) from the aluminium pouch and place the tubes in an appropriate holder.
- Add the following to each tube:
a) Sterile water
b) Forward primer
c) Reverse primer
d) Probe or intercalating dye
e) Optional: Optimal amount of MgCl2 for specific reaction - Close the caps and mix the contents of each tube by tapping them gently with your finger.
- Transfer tubes to template addition lab and proceed with the addition of template.
f) DNA template*
Total reaction volume for all components (a–f) = 25 μL
*Template titrations prepared by making serial dilutions
Thermal cycling conditions
Different systems require different thermal cycling conditions. Conditions for some common systems follow:
TaqMan™ probes:
95°C for 5 min
Three-step cycling (35–40 cycles)
95°C for 30 s
55°C for 30 s
72°C for 60 s (DETECTION step)
MGB™ Eclipse probe system:
50°C for 2 min
95°C for 30 s
Three-step cycling (40–50 cycles)
95°C for 10 s
56°C for 30 s (DETECTION step)
76°C for 30 s
Melt-curve analysis: ABI PRISM 7900 manufacturer’s recommended default settings
GelStar™ nucleic acid gel stain:
95°C for 5 min
Three-step cycling (35–45 cycles)
95°C for 30 s
55°C for 30 s
72°C for 60 s
Melt-curve analysis: Smart Cycler manufacturer’s recommended default settings
Results
Low background
Nonspecific background amplification can interfere with desired PCR results. Background amplification reduces primer and nucleotides for product formation, thereby decreasing overall PCR efficiency. In addition, in real-time applications that rely on intercalating dyes to detect products, background amplification can create unacceptable levels of background fluorescence.
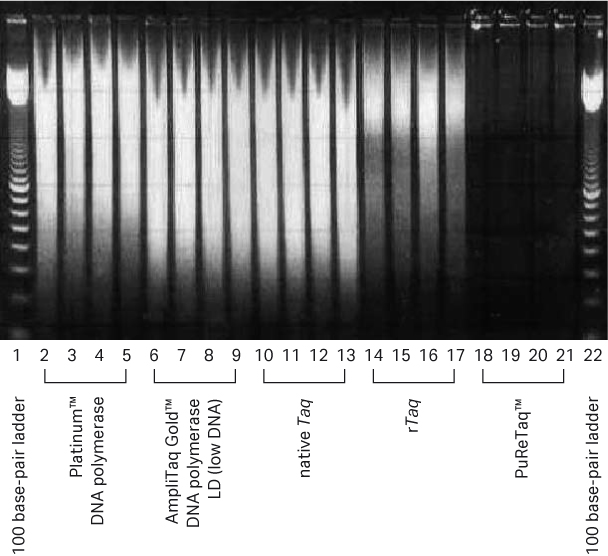
To demonstrate the reduction of nonspecific background amplification in the Ready-To-Go™ PCR Beads, an endpoint PCR assay and random hexamers were used to compare bead results to those obtained using other commercially available Taq polymerases. Nonspecific background amplification resulting from residual nucleic acid contamination was visualized as a broad smear of products on agarose gels. Recombinant Taq polymerase from Cytiva showed low amounts of product on agarose gel compared to the two competitors’ Taq polymerases, but the subsequent treatment to produce PuReTaq™ eliminated the residual general background observed with all the other systems (Fig 1).
Fig 1. Comparative enzyme performance evaluation using random primers. From left to right, each well contains 10 μL from an original 25 μL reaction volume. Five commercially available enzymes were compared using random hexamers with no template added. All reactions were run in quadruplicate.
Real-time reaction performance
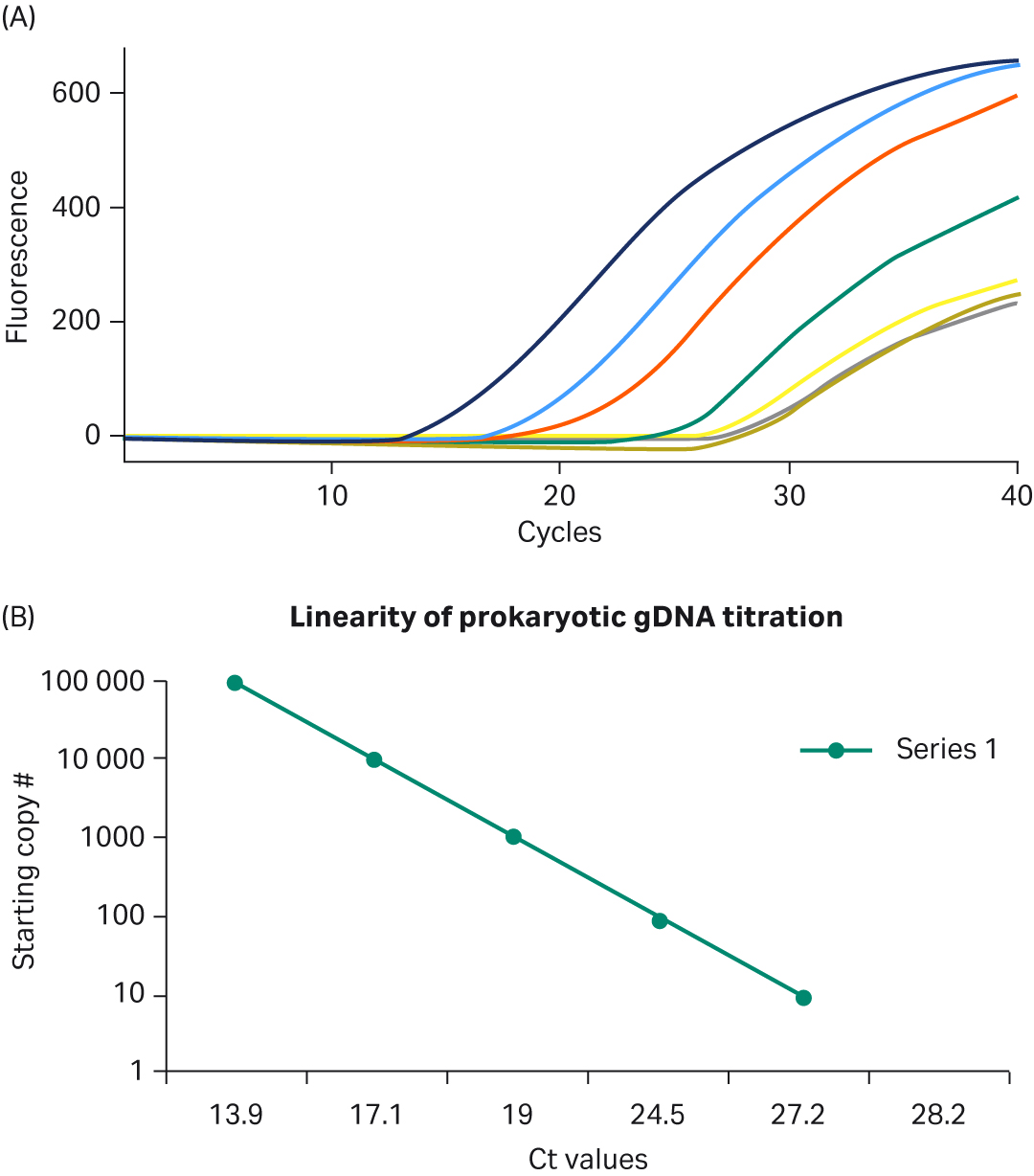
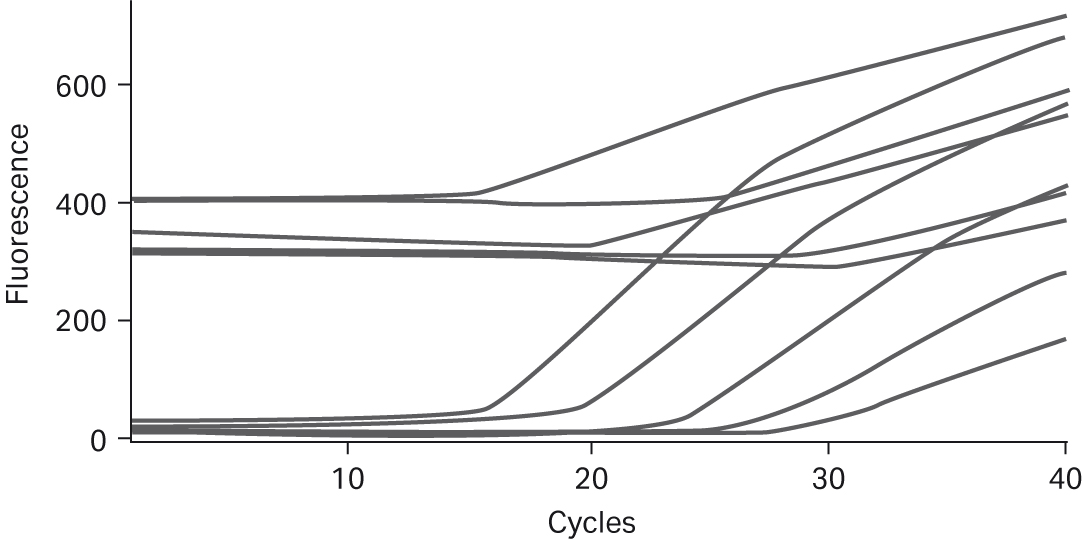
To assess the performance of PCR samples conducted with the Ready-To-Go™ PCR Beads, real-time PCR was run using TaqMan™ probes and human DNA template. As shown in Figure 2A, the beads produced robust growth curves and low background fluorescence. In addition, as shown in Figure 2B, template titrations were linear after optimizing Mg2+ and cycling conditions; no other additions to the PuReTaq™ bead were necessary to optimize these representative reactions. The results shown here were obtained by running reactions on the Smart Cycler™, but comparable performance was observed when tests were completed on the ABI PRISM™ 7700 and 7900 Sequence Detection Systems or on the Roto-Gene 3000™ (data not shown). The beads used in these assays were stored at room temperature according to the manufacturer’s instructions, and no drop-off in performance was observed when the same tests, under the same conditions, were run over one year later.
Fig 2. A. Top five real-time PCR growth curves obtained for prokaryotic template and no-template controls. Template titrations each contain one Ready-To-Go™ PCR Bead; 105 to 10 copies of genomic DNA from E.coli DH1λ; 20 pmol 16s rRNA primers (forward and reverse); 5 pmol of 16s rRNA FAM-TAMRA probe; 6.1 mM MgCl2; and sterile H2O. Reactions were amplified as follows: 95°C for 300 s, then 95°C for 30 s, 55°C for 30 s, and 72°C for 60 s for 40 cycles. B. Linearity of prokaryotic gDNA template titration using TaqMan™ primers/probe. B. Linearity of prokaryotic gDNA template titration using TaqMan™ primers/probe
Reproducibility
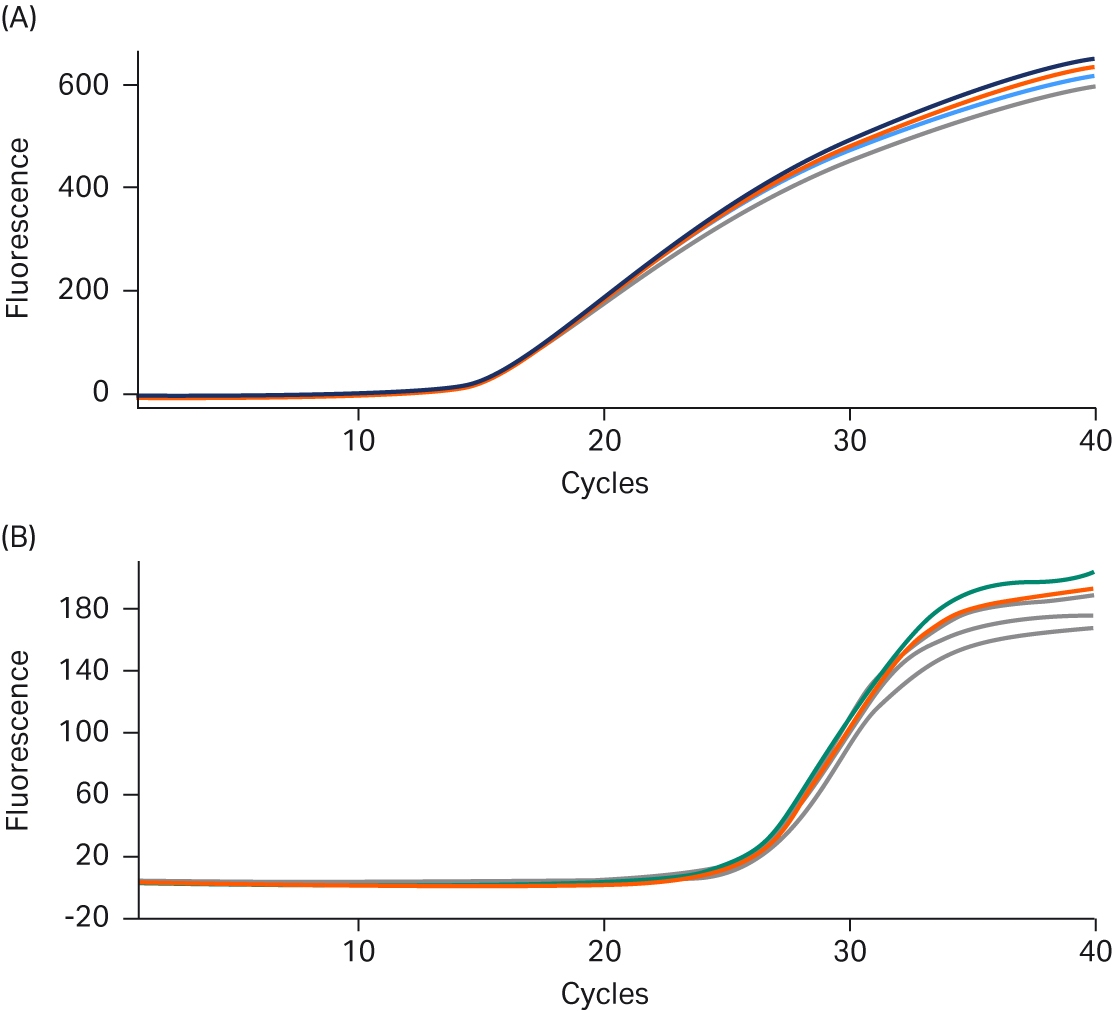
Reproducibility of the new Ready-To-Go™ PCR Beads was assessed bead-to-bead and lot-to-lot. Figures 3A and 3B show replicate real-time results from TaqMan™ assays and intercalating-dye-based assays, respectively. Eleven replicates were run for each assay type, without any amplitude normalization: thresholds were calculated with the Smart Cycler™ at three standard deviations above baseline signal. The fluorescence threshold (Ct) mean for the TaqMan™ growth curves was 14.49 cycles, the standard deviation was 0.062 cycles, and %CV was 0.41% (Fig 3A). The threshold mean, standard deviation, and %CV for the GelStar™ nucleic acid gel stain curves were 25.98, 0.24, and 0.92%, respectively (Fig 3B). These results confirm a high degree of reproducibility between beads at a fixed template input amount.
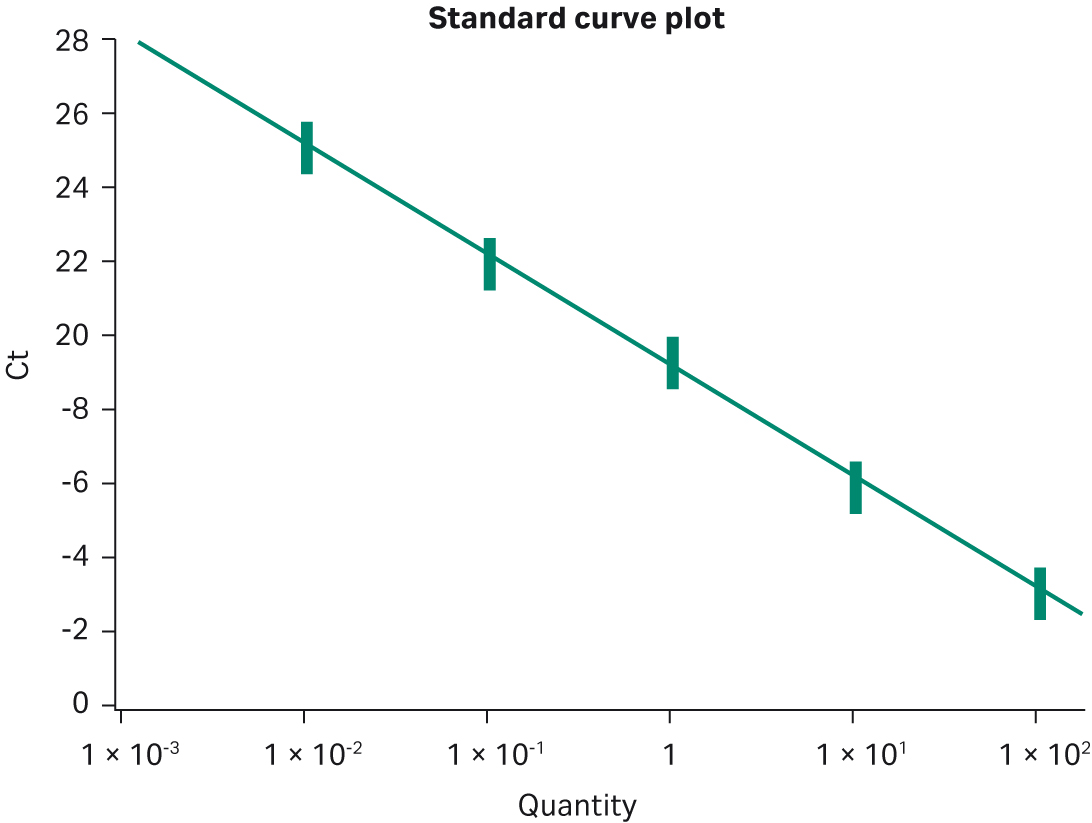
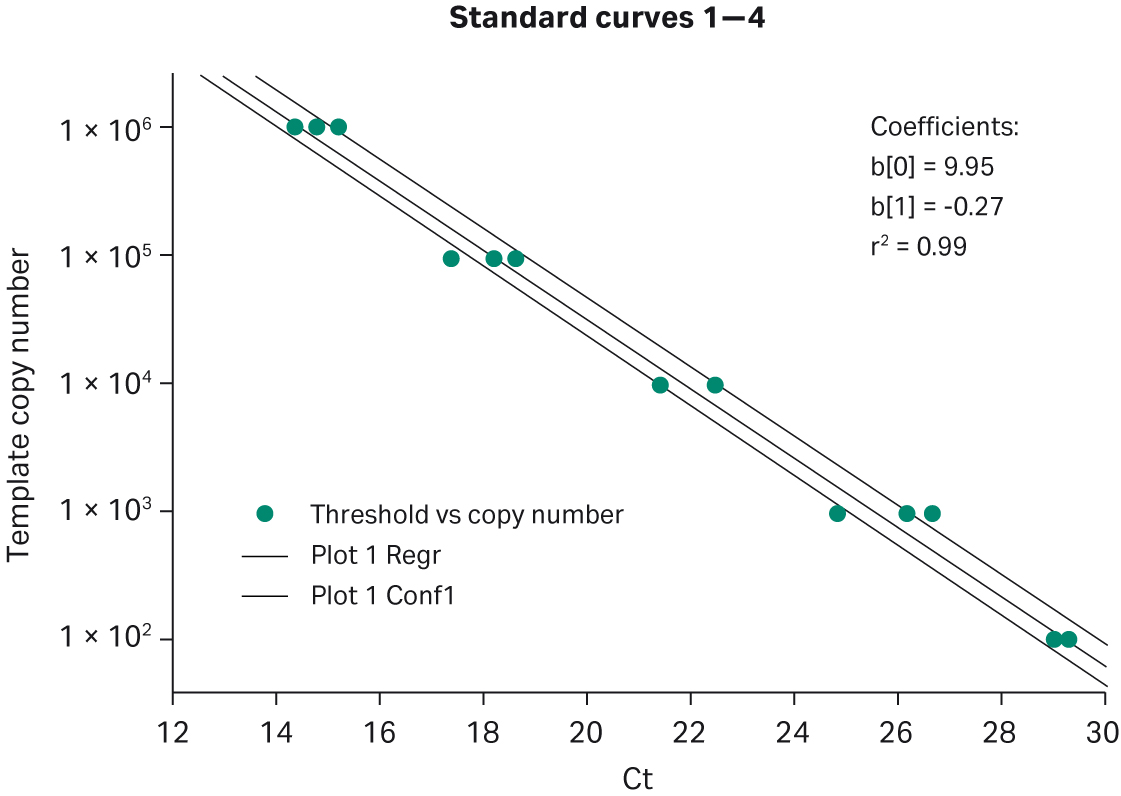
Multiple template titrations were also run with the MGB™ Eclipse Probe System real-time PCR system. This system differs from either intercalating dye or TaqMan™ assays in that it generates maximum fluorescent signal upon annealing to the PCR amplicon during the anneal phase of the PCR cycle. Template titrations over 5 logs (0.01 ng–100 ng) produced standard deviations of 0.25, 0.24, 0.31, 0.21, and 0.21 cycles and %CVs between 1 and 1.6% (Fig 4). These data provide strong evidence of the bead-to-bead reproducibility and broad applicability of Ready-To-Go™ PCR Beads across different real-time technologies. In the final reproducibility test, four manufactured lots of beads were compared in a series of template titrations using intercalating dye. As shown in Figure 5 and Table 1, the standard deviations of cycle thresholds were 0.12, 0.45, 0.45, and 0.3 for each copy number tested, with %CV values between 0.41 and 2%. These data indicate that customers can rely on lot-to-lot as well as bead-to-bead reproducibility.
Fig 3. A. TaqMan™ assay reproducibility. Real-time PCR data highlighting reproducibility for 11 replicates of 107 copies of E.coli (DH1λ+) genomic DNA. B. Intercalating dye assay reproducibility. Real-time PCR data highlighting reproducibility for 11 replicates of 50 pg of Lambda DNA.
Fig 4. Bead-to-bead reproducibility using MGB™ Eclipse Probe System. Twelve replicates of a five-concentration titration were performed. BAC-containing human DNA sequence was used as template from 100 ng to 10 pg. Optimal Mg2+ was 5 mM. R2 = 0.99.
Fig 5. Lot-to-lot reproducibility. This graph shows four different template titrations, each performed with a separate lot of Ready-To-Go™ PCR Beads. Lambda DNA titration from 106 to 102 copies, detected with intercalating dyes.
Table 1. Lot-to-lot reproducibility. Four different lots of manufactured Ready-To-Go™ PCR Beads were chosen randomly and used for intercalating-dye-based assays. The number of copies of Lambda DNA template are listed at the top of table.
| Amount of template | 1E+05c | 1E+04c | 1E+03c | 1E+02c |
| lot A | 14.75 | 17.38 | 21.39 | 28.98 |
| lot B | 15.21 | 18.15 | 21.31 | 28.92 |
| lot C | 14.76 | 18.20 | 21.38 | 29.22 |
| lot D | 14.35 | 1.64 | 22.40 | 29.16 |
| Mean | 14.77 | 18.09 | 21.62 | 29.07 |
| STD Deviation | 0.30 | 0.45 | 0.45 | 0.12 |
| %CV | 2.00 | 2.50 | 2.10 | 0.41 |
Flexibility; Mg2+ concentration
While scientists demand well-designed, highly reproducible material for performing PCR, they also require flexibility in optimizing their particular amplification while minimizing background fluorescence. The new Ready-To-Go™ PCR Beads have been intentionally formulated with a low concentration of Mg2+, making it easy to increase the level to optimize reaction performance. Figure 6 shows how higher concentrations of Mg2+ can affect background fluorescence: a four-fold change in Mg2+ leads to a three-fold decrease in background fluorescence. This ability to control Mg2+ concentration can be exploited to optimize the signal-to-background ratio and can improve the lower limits of detection and reproducibility of sequence detection at low copy numbers.
Fig 6. TaqMan™ assay to detect prokaryotic sequences. All curves are shown without background subtraction on a Smart Cycler™. MgCl2 concentration for the curves with higher background is 1.5 mM and lower background is 6.1 mM. Template levels were 105 copies to 10 copies.
Competitive performance
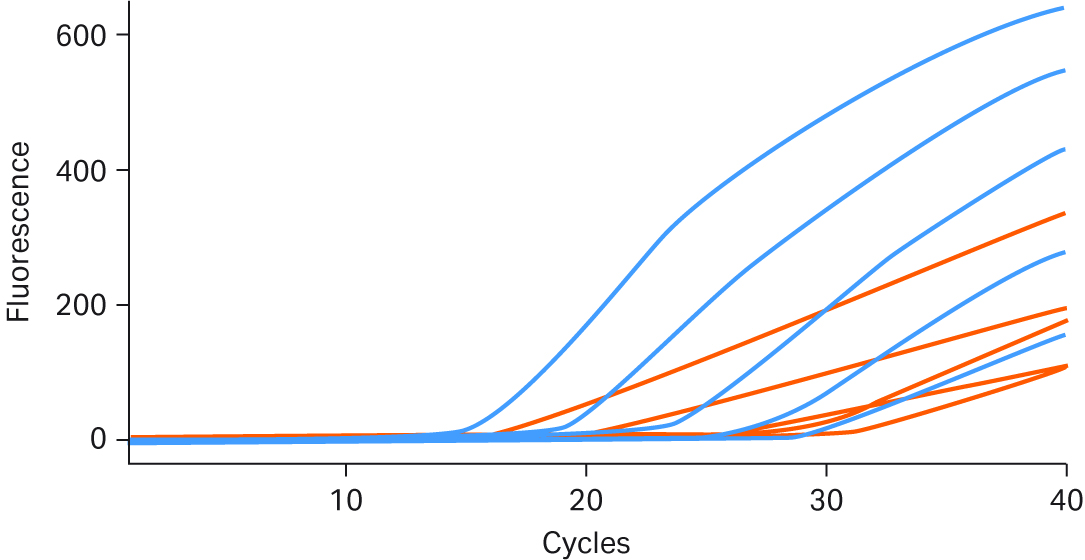
Real-time performance of Ready-To-Go™ PCR Beads was compared to the AmpliTaq™ Gold PCR Master Mix run using the same assay, template, and instrument. Both products were stored under the manufacturer’s recommended conditions and run using optimum assay conditions. As Figure 7 shows, the Ready-To-Go™ Beads yielded more robust growth curves in amplitude. Even though the competitor’s product gave positive results for each template copy number tested, its signal amplitude was lower, making low-end detection ambiguous and offering questionable reproducibility.
Fig 7. The dark blue real-time PCR growth curves are results obtained using Ready-To-Go™ PCR Beads; red growth curves are for AmpliTaq™ Gold PCR Master Mix. All curves were obtained under identical conditions, with E.coli DH1λ+ genomic DNA titrated from 105 to 10 copies.
Results summary
The results discussed in this application note demonstrate that Ready-To-Go™ PCR Beads
- Yield robust, complete, stable reactions with low-levels of background contaminating DNA in real-time PCR performed with intercalating dyes, TaqMan™ assays, or MGB™ Eclipse Probe System.
- Offer exceptional reproducibility bead-to-bead and lot-to-lot, even across multiple systems.
- Contain a low Mg2+ concentration that can be used as is or titrated to optimize signal-to-background for each reaction.
- Outperform competitors’ PCR premixes in side-by-side comparisons using the same assays, templates, and instruments.