GenomiPhi™ V3 Ready-To-Go™ DNA Amplification Kit is an ambient-temperature-stable version of GenomiPhi™ for whole genome amplification (WGA) designed to increase yield, simplify product handling and storage, and reduce the risk of contamination.
We have produced an ambient-temperature-stable version of GenomiPhi™ for whole genome amplification (WGA) called GenomiPhi™ V3 Ready-To-Go™ DNA Amplification Kit to increase yield, simplify product handling and storage, improve consistency, and reduce the risk of contamination. For comparative performance analysis, we amplified human genomic DNA (gDNA) using GenomiPhi™ V3 Ready-To-Go™ and GenomiPhi™ V2 DNA amplification kits. The results show that the GenomiPhi™ V3 Ready-To-Go™ Amplification Kit produced greater yields than GenomiPhi™ V2 DNA Amplification Kit. We used WGA products from both kits in downstream applications such as endpoint PCR and STR analysis.
GenomiPhi™ technology uses Phi29 DNA polymerase and random hexamer primers for isothermal WGA. GenomiPhi™ amplification reactions can generate microgram quantities of high molecular weight DNA from nanogram quantities of starting material. Phi29 DNA polymerase produces high fidelity during DNA replication due to its proofreading 3’–5’ exonuclease activity (1, 2). We designed GenomiPhi™ V3 Ready-To-Go™ DNA Amplification Kit, shown in Figure 1, to offer you a simplified workflow plus long-term ambient-temperature stability in a Ready-To-Go™ format. As a result of these improvements, GenomiPhi™ V3 Ready-To-Go™ Amplification Kit outperforms GenomiPhi™ V2 DNA Amplification Kit in WGA reactions.
Fig 1. GenomiPhi™ Ready-To-Go™ DNA amplification kit in convenient ambient temperature stable format provides reproducible and reliable performance for demanding applications such as whole genome sequencing, genotyping, and array CGH.
Find out more about our GenomiPhi™ DNA Amplification Kits by reading our datafileMaterials and methods
GenomiPhi™ amplification
Purified human gDNA (10 ng) and lambda DNA (10 ng) were amplified using the standard protocols for each of the two GenomiPhi™ kits (Tables 1 and 2). DNA yield from each amplification reaction was quantitated using Quant-IT™ PicoGreen™ dsDNA quantitation reagent. The samples were also taken at 30 min intervals and quantitated with PicoGreen™ assay to monitor amplification yield throughout the recommended 90 min incubation period and beyond.
Table 1. Overview of standard protocols for GenomiPhi™ V2 DNA Amplification Kit
| GenomiPhi™ V2 DNA Amplification Kit | |
| Remove from freezer and thaw on ice | |
| Input DNA (10 ng) | 1 μL |
| Sample buffer | 9 μL |
| Heat to 95°C 3 min, cool to 4°C on ice | |
| Reaction to buffer | 9 μL |
| Enzyme mix | 1 μL |
| Incubate at 30°C | 90 min |
| Inactivate enzyme at 65°C for 10 min | |
| Typical product yield | 4 to 7 μg |
Table 2. Overview of standard protocols for GenomiPhi™ V3 Ready-To-Go™ DNA Amplification Kit
| GenomiPhi™ V3 Ready-To-Go™ DNA Amplification Kit | |
| Remove from shelf | |
| Input DNA (10 ng) | 1 μL |
| PCR-grade water | 9 μL |
| 2x denaturation buffer | 10 μL |
| Heat to 95°C 3 min, cool to 4°C on ice | |
| Add sample to cake | 20 μL |
| Incubate at 30°C | 90 min |
| Inactivate enzyme at 65°C for 10 min | |
| Typical product yield | 12 to 20 μg |
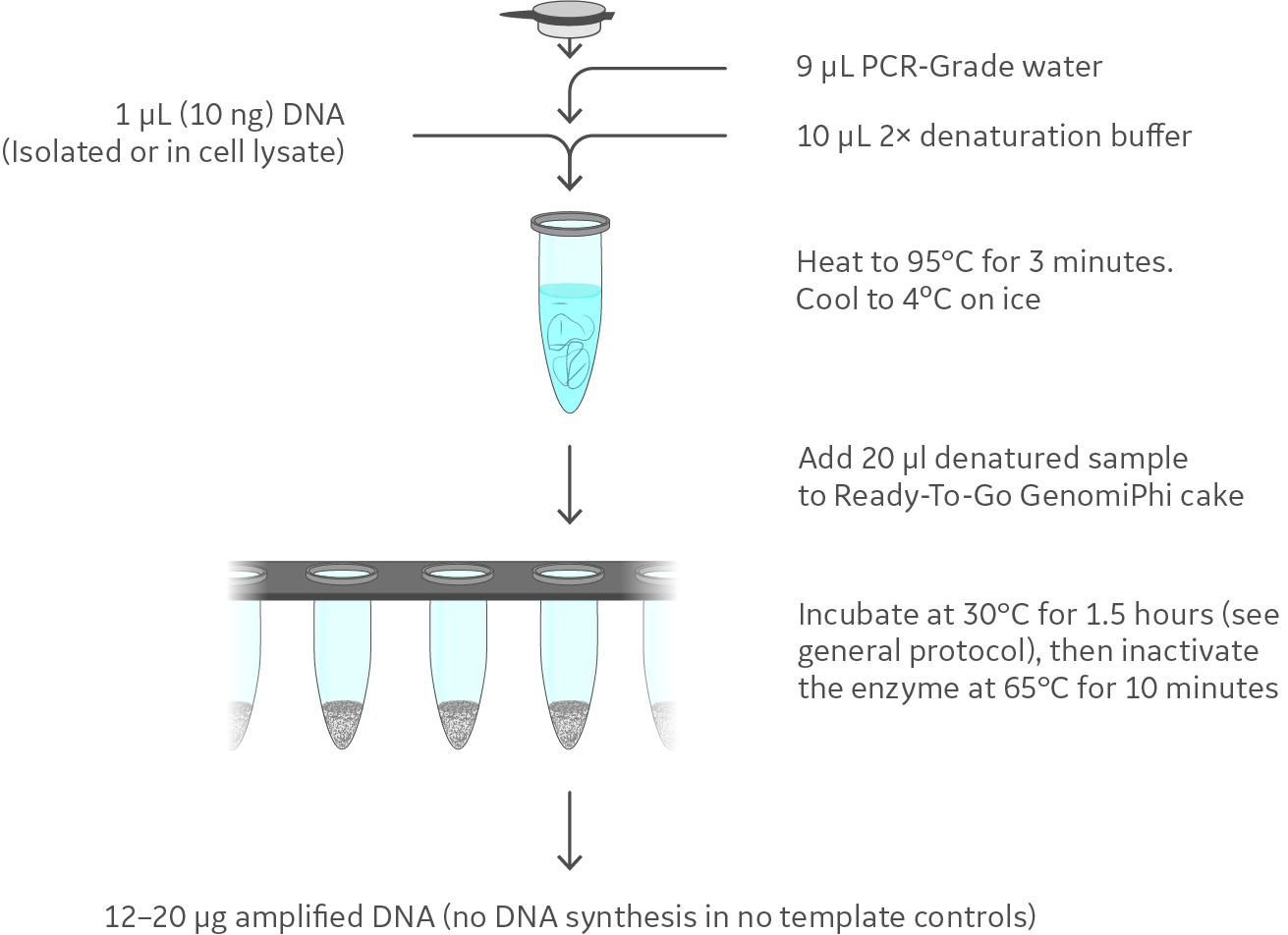
Negative amplifications [i.e., no template control (NTC)] in which template DNA was substituted with water were carried out in parallel. A schematic representation of the GenomiPhi™ V3 Ready-To-Go™ DNA Amplification Kit workflow is shown in Figure 2. Amplified human gDNA was carried through to STR analysis.
Fig 2. The workflow for GenomiPhi™ V3 Ready-To-Go™ DNA Amplification Kit
Short tandem repeat (STR) genotyping
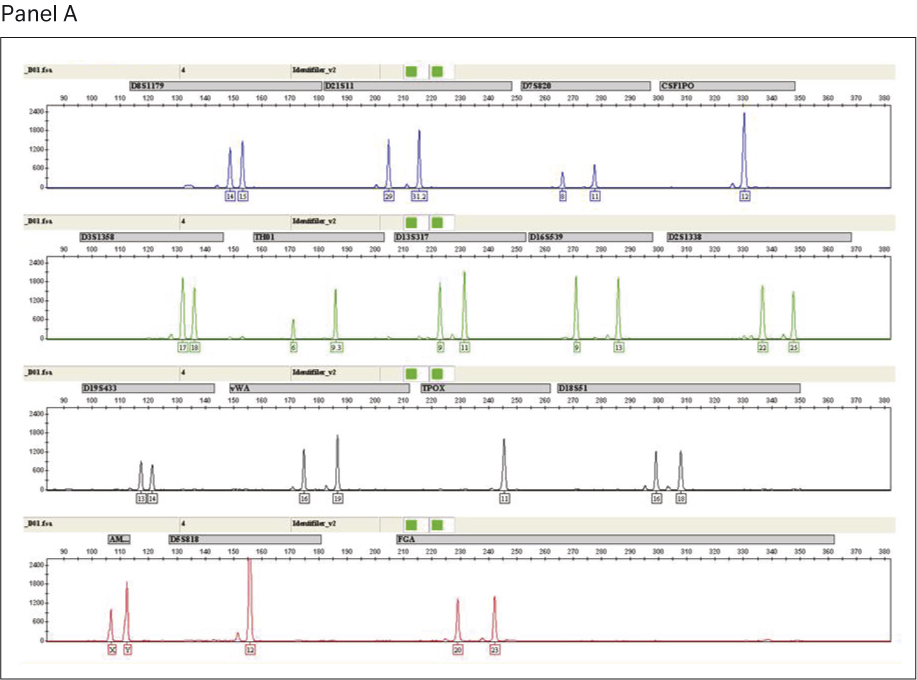
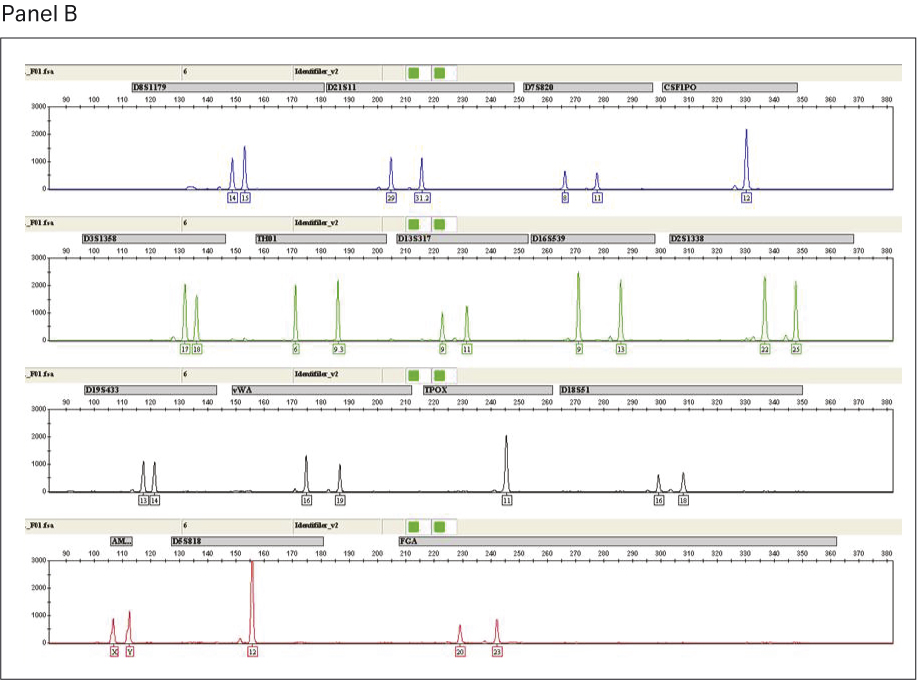
STR genotyping was performed to assess the quality and purity of amplified DNA generated from single-source human gDNA using GenomiPhi™ V3 Ready-To-Go™ and GenomiPhi™ V2 DNA amplification kits. A total of 15 loci (plus amelogenin) were investigated for each sample using the AmpFLSTR™ Identifiler™ PCR Amplification Kit according to the manufacturer’s protocol. STR amplifications were performed using a GeneAmp™ PCR system 9700. Capillary electrophoresis was performed on a 3130xl genetic analyser, followed by analysis using GeneMapper™ ID v3.2 software.
Endpoint PCR analysis
Endpoint PCR amplification of the human p53 gene was performed on amplified human gDNA and corresponding WGA negative control reactions from both GenomiPhi™ kits. Each GenomiPhi™ amplification reaction was carried out according to the standard protocol using 10 ng of input DNA. The products were quantitated and diluted prior to PCR amplification. Each PCR was prepared using two PureTaq™ Ready-To-Go™ PCR beads, 1 μL each of 10 μM forward and reverse p53 primers, 4 ng of DNA, plus sterile distilled water to a total volume of 50 μL. PCR amplifications were performed using a PTC-200 thermal cycler (MJ Research) with the following PCR cycling conditions: 95°C for 3 min, followed by 35 cycles of 95°C for 30 s; 55°C for 1 min; 72°C for 1 min.
2. Results and discussion
GenomiPhi™ DNA yield
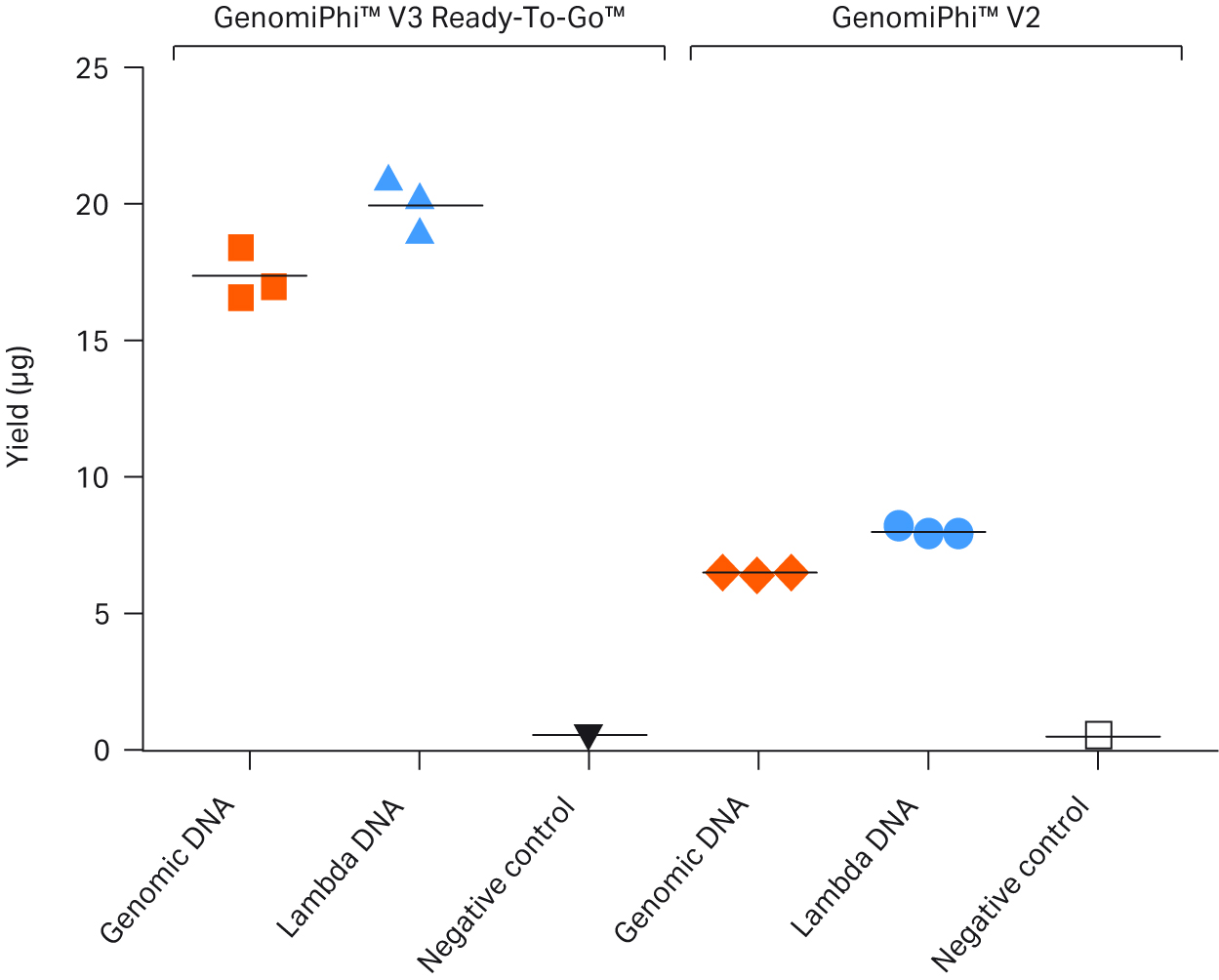
DNA quantitation after amplification of human gDNA or Lambda DNA using GenomiPhi™ V3 Ready-To-Go™ DNA Amplification Kit showed a significantly increased yield compared to GenomiPhi™ V2 DNA Amplification Kit for both human genomic and lambda DNA (Fig 3).
Fig 3. Determination of the yield of human gDNA or lambda DNA amplified with either GenomiPhi™ V3 Ready-To-Go™ or GenomiPhi™ V2 DNA amplification kits.
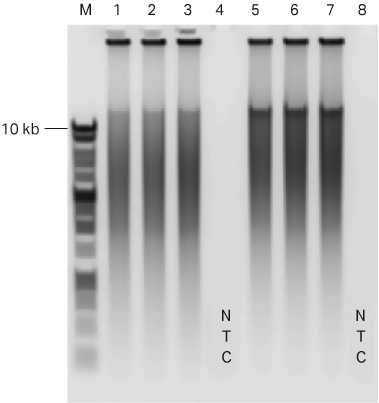
The negative control reactions did not show any DNA synthesis during the 90 min incubation period (Figures 3 and 4) for both kits. Agarose gel analysis of amplification reactions confirmed the presence of high molecular weight DNA in all the reactions containing template DNA (Fig 4).
Fig 4. Agarose gel (1% agarose, 1x TAE) showing the amplification product of human gDNA (10 ng) in GenomiPhi™ V2 (lanes 1 to 3) or GenomiPhi™ V3 Ready-To-Go™ kit (lanes 5 to 7). NTC represents no template controls for GenomiPhi™ V2 (lane 4) or GenomiPhi™ V3 Ready-To-Go™ kit (lane 8).
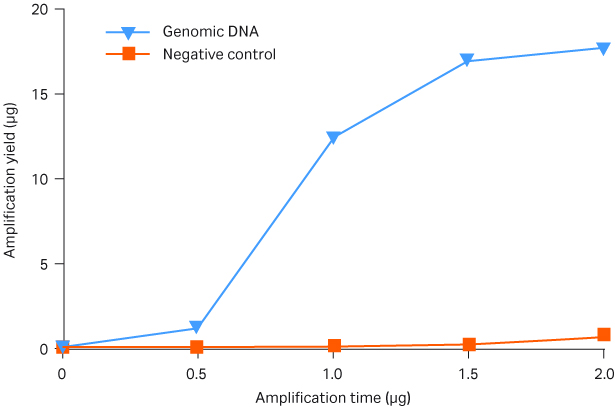
The yield from GenomiPhi™ V3 Ready-To-Go™ reactions was determined at 30 min intervals using a PicoGreen™ assay. The amplification reaction reached a plateau at the end of the recommended 90 min incubation period without any appreciable background material in the negative controls (Fig 5).
Fig 5. Time course of gDNA (10 ng) amplification with GenomiPhi™ V3 Ready-To-Go™ Amplification Kit. Water was substituted for template DNA in the negative control reactions.
STR analysis
STR analysis produced the expected allele calls for the full profile of 15 loci (plus amelogenin) for the non-amplified single-source gDNA control. The profiles for gDNA amplified using GenomiPhi™ V2 and the GenomiPhi™ V3 Ready-To-Go™ DNA amplification kits were comparable (Fig 6); both products generated the same allele calls as the gDNA control (Table 3). In summary, we obtained 100% accuracy in allele calls from a single-source gDNA as starting material after amplification with GenomiPhi™ V3 Ready-To-Go™ and GenomiPhi™ V2 DNA amplification kits.
Table 3. Allele calls for 16 loci determined by STR profiling of DNA (GenomiPhi™ V2 or V3-amplified and non-amplified control gDNA).
| GenomiPhi™ V2 DNA Amplification Kit | GenomiPhi™ V3 Ready-To-Go™ DNA Amplification Kit | Control gDNA (Not amplified) | ||||
| Marker | Allele 1 | Allele 2 | Allele 1 | Allele 2 | Allele 1 | Allele 2 |
| D8S1179 | 14 | 15 | 14 | 15 | 14 | 15 |
| D21S11 | 29 | 31.2 | 29 | 31.2 | 29 | 31.2 |
| D7S820 | 8 | 11 | 8 | 11 | 8 | 11 |
| CSF1PO | 12 | - | 12 | - | 12 | - |
| D3S1358 | 17 | 18 | 17 | 18 | 17 | 18 |
| TH01 | 6 | 9.3 | 6 | 9.3 | 6 | 9.3 |
| D13S317 | 9 | 11 | 9 | 11 | 9 | 11 |
| D16S539 | 9 | 13 | 9 | 13 | 9 | 13 |
| D2S1338 | 22 | 25 | 22 | 25 | 22 | 25 |
| D19S433 | 13 | 14 | 13 | 14 | 13 | 14 |
| vWA | 16 | 19 | 16 | 19 | 16 | 19 |
| TPOX | 11 | - | 11 | - | 11 | - |
| D18S51 | 16 | 18 | 16 | 18 | 16 | 18 |
| AMEL | X | Y | X | Y | X | Y |
| D5S818 | 12 | - | 12 | - | 12 | - |
| FGA | 20 | 23 | 20 | 23 | 20 | 23 |
Fig 6. Output from GeneMapper™ software showing alleles from the 15 AmpFLSTR™ Identifiler loci for GenomiPhi™ V2 WGA (panel A) and GenomiPhi™ V3 Ready-To-Go™ WGA (Panel B).
PCR analysis
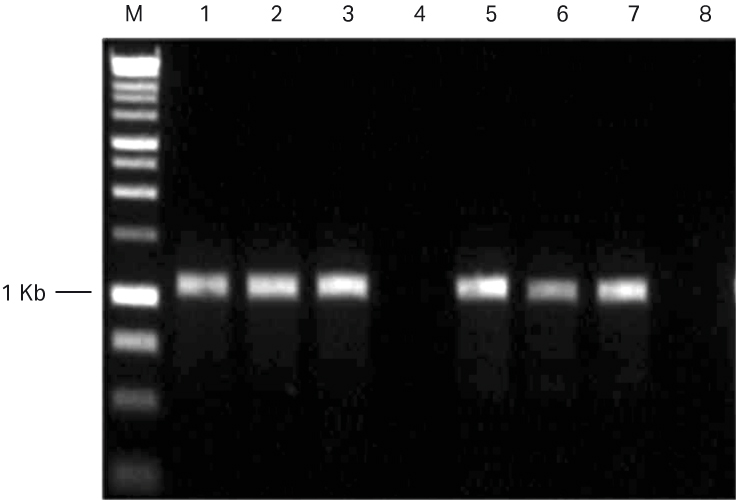
The expected 1.1 kb p53 PCR product was amplified from both GenomiPhi™ V2 and GenomiPhi™ V3 Ready-To-Go™ kit WGA samples (Fig 7). No product was generated in the WGA negative (no template) control reactions.
Fig 7. Agarose gel (1% agarose, 1x TAE) showing the expected 1.1 kb p53 PCR product for GenomiPhi™ V2 WGA reactions (lanes 1 to 3) and GenomiPhi™ V3 Ready-To-Go™ WGA reactions (lanes 5 to 7). WGA negative (no template) controls for GenomiPhi™ V2 and GenomiPhi™ V3 Ready-To-Go™ showed no PCR product (lanes 4 and 8), M = 1 kb ladder.
Find out more about using the GenomiPhi™ DNA Amplifications Kits3. Conclusions
The incorporation of a Ready-To-Go™ format into the design of GenomiPhi™ V3 Ready-To-Go™ DNA Amplification Kit has resulted in several advantages with respect to workflow and handling, and a significant reduction in the overall process time. The drawback of GenomiPhi™ V2 kits, and indeed other DNA amplification kits in liquid format, is the need to keep them frozen to maintain performance. Apart from the inconvenience of freezer storage, the reagents must be thawed carefully on ice, mixed for use, and returned promptly to storage. The ambient temperature stability of GenomiPhi™ V3 Ready-To-Go™ technology removes these requirements. The addition of denatured DNA directly to the stabilized reaction mix of GenomiPhi™ V3 Ready-To-Go™ DNA Amplification Kit initiates the amplification reaction, thus eliminating the setup stage involved with the GenomiPhi™ V2 DNA Amplification Kit, in which the polymerase is exposed in the absence of a template.
In summary:
- We have used GenomiPhi™ V2 and GenomiPhi™ V3 Ready-To-Go™ DNA amplification kits to successfully amplify human gDNA for typical downstream applications.
- Using PCR primers specific for the p53 tumour suppressor and the amplified product from GenomiPhi™ V2 and GenomiPhi™ V3 Ready-To-Go™ DNA amplification kits as templates, we obtained the expected 1.1 Kb product in a PCR.
- DNA amplified with GenomiPhi™ V2 and GenomiPhi™ V3 Ready-To-Go™ DNA amplification kits provided 100% identical allele calls to a single-source human gDNA that had not been previously amplified in STR genotyping.
- While the DNA products from GenomiPhi™ V2 and GenomiPhi™ V3 Ready-To-Go™ DNA amplification kits produced similar results in downstream applications, the yield from GenomiPhi™ V3 Ready-To-Go™ was significantly greater.
Read our knowledge article on: Trends in rolling circle amplification to find out how TempliPhi amplification kits use isothermal rolling circle amplification (RCA), enabled by phi29 DNA polymerase, to support advancements with manipulation, error-free production and assembly of nucleic acids.
- Estaban, J.A. et al., Fidelity of phi29 DNA polymerase, J. Biol. Chem. 268, 2719–2726 (1993).
- Nelson, J.R. et al; TempliPhi, Phi29 DNA polymerase based rolling circle amplification of templates for DNA Sequencing, BioTechniques 32, S44-S47 (2002).