Rapid and accurate detection of viral pathogens is critical to the treatment, monitoring, successful management and control of epidemics. Nucleic acid amplification tests (NAAT) are a reliable, sensitive, and accurate diagnostic approach used in viral detection (1,2). This approach involves the isolation of nucleic acids from a sample, which is not only time and resource consuming but increases the chances of accidental contamination and human error.
In a period of high demand, such as the current COVID-19 pandemic, a shortage of nucleic acid isolation kits can exacerbate the limitations of the nucleic acids-dependent viral detection methods. The high demand for sample testing makes it difficult for diagnostic testing laboratories to provide accurate results in a timely manner.
Direct reverse transcriptome polymerase chain reaction (RT-PCR) from unpurified samples promises to reduce the time and resources required for sample testing by eliminating the need for the isolation of nucleic acids. However, several technical limitations, including sensitivity due to the presence of RT-PCR inhibitors and nucleic acid degradation in the sample, must be addressed to make this approach a viable alternative. At Cytiva, we have developed a proprietary chemistry—anti-inhibitor complex (AIC)—that addresses these limitations. In this proof of concept, we show that the chemistry allows for an extraction-free NAAT, thereby significantly reducing the time and resources required to detect viral particles in nasopharyngeal swabs collected in transport medium (Fig 1). The proprietary formulation provides a robust chemistry where low abundance nucleic acids such as viral RNAs in the sample are stable. The formulation also enhances the assay sensitivity by increasing the tolerance of the RT-PCR enzymes to inhibitors present in the samples.
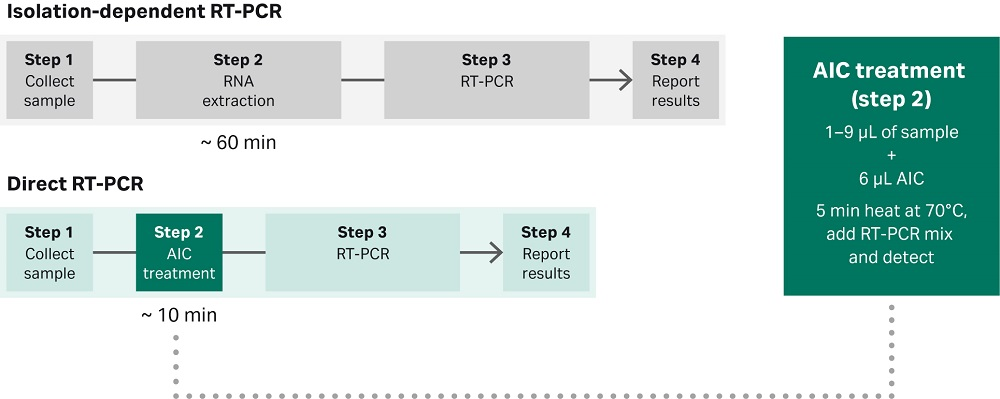
Fig 1. The Cytiva RT-PCR anti-inhibitor complex (AIC) allows direct RT-PCR detection of nucleic acid material from unprocessed samples. The proprietary RT-PCR AIC eliminates the need for nucleic acid isolation, requiring only a short five-minute heating treatment at 70°C prior the RT-PCR reaction.
Results
By combining the proprietary Cytiva AIC formulation with one-step reverse transcription and DNA amplification (RT-PCR) kits, we have increased the sensitivity, reproducibility, and robustness of the assay for detection of viral particles in unpurified samples. As shown in Figure 2: A-B, the Cytiva AIC chemistry allows the detection of viral RNA directly from unpurified nasopharyngeal samples spiked in with as little as 10 copies of viral particles per reaction, without the need for nucleic acid isolation, reducing both costs and the time to obtain a result.
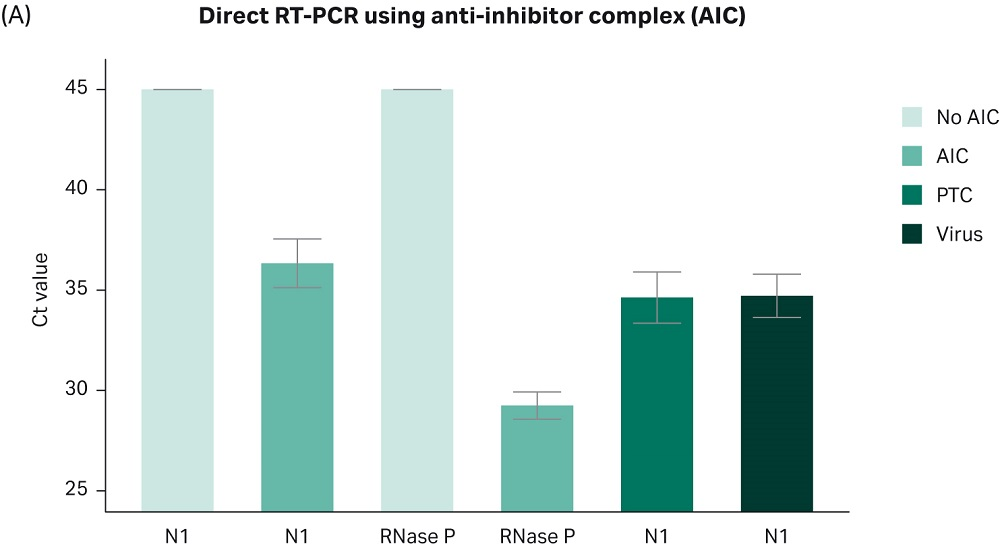
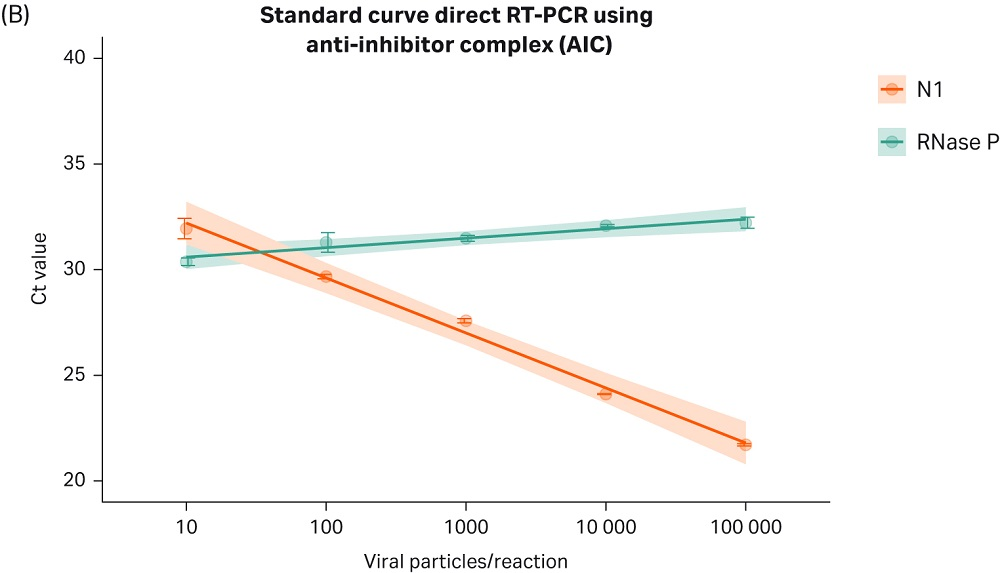
Fig 2. The Cytiva RT-PCR AIC mix provides robust, sensitive and accurate detection of COVID-19 viral particles directly from unpurified patient samples. Human nasopharyngeal swabs collected in transport medium (Nasopharyngeal swabs from patients tested negative for COVID-19, Tissue Solutions, FM Reference: QQPE7) were spiked in with varying amounts of heat inactivated SARS-CoV-2 viral particles (SeraCare™, Cat. N.0505-0126, Lot N.10481654; or ATCC VR-1986HK). 5 µL of sample were mixed with 6 µL of AIC reagent, heated for 5 minutes at 70°C, and subsequently combined with primers (IDT Primes, N1 or RNase P, 2019-nCoV RUO Kit: Integrated DNA Technologies™, Cat.N. 10006713), and RT-PCR master mix. The reverse transcription and amplification reactions were carried out in one step, following the kit manufacturer’s recommendations (TaqPath™ 1-step Multiplex Master Mix for RT-PCR, Applied Biosytems™, Cat. N. A28525). (A) Bar plot shows Ct values obtained by real-time RT-PCR using primer set N1 targeting the N gene of SARS-CoV-2 virus. The plot shows the results combined from three independent experiments ± SD, with three technical replicates run each time. As positive control primers targeting the human RNase P gene, which amplifies genomic material from human cells was used. Plasmid Template Control (PTC) (20 cp/reaction): Plasmid coding for same artificial sequence contained in viral particles. Virus (10 cp/reaction): Artificial viral containing a portion of the SARS-CoV-2 viral genome. (B) Real-time RT-PCR standard curve demonstrates linear and sensitive detection of viral RNA across a wide range of viral loads. Experiment was carried out as described above. Viral load ranged from 10-10e5 particles per reaction.
With COVID-19 NAAT testing, nasopharyngeal swabs are the recommended sample type from patients. These swabs are commonly stored in transport medium, which can itself be an inhibitor for the amplification reaction. Nonetheless, commercial RT-PCR kits currently in the market have limited tolerance to transport media. This tolerance can be up to 25% of the reaction volume (3, 4) and could potentially limit an assay performance, especially when working with low viral load samples such as those observed in some COVID-19 patients, where viral loads vary with the progression of the disease (5,6). Hence, a wider range of tolerance to transport media may be advantageous for a successful detection of low viral loads. One way to address this limitation is by combining an optimized one-step RT-PCR master mix formulation with the RT-PCR anti-inhibitor complex. The resulting Cytiva direct RT-PCR chemistry exhibits high tolerance to transport media by being capable of detecting low viral loads even when the sample-transport media volume reaches 45% of the final reaction volume (Fig 3). This can be beneficial when testing border-line negative/positive samples, by doubling the sample volume tested without affecting assay sensitivity.
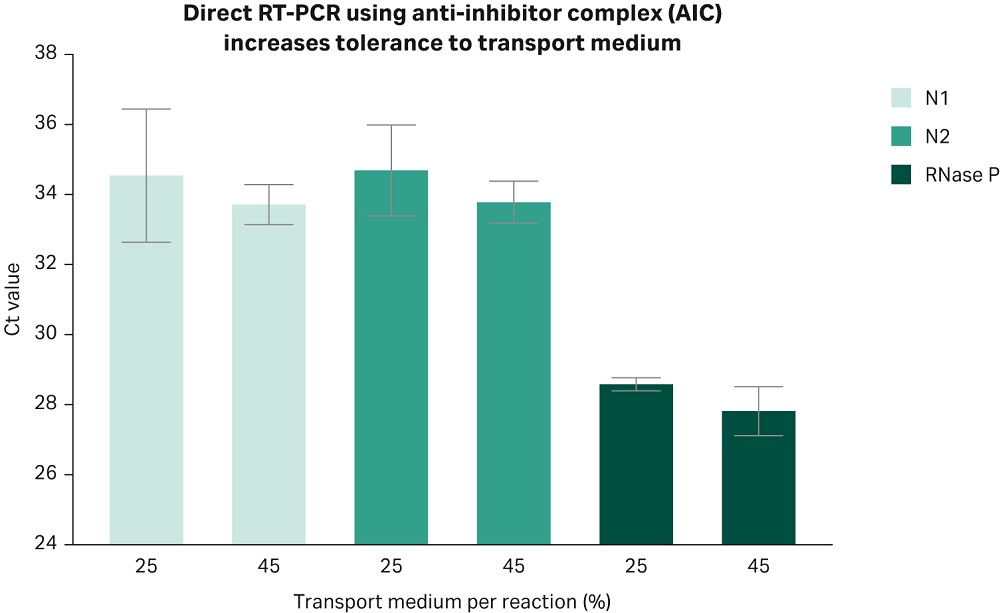
Fig 3. The Cytiva direct RT-PCR chemistry is tolerant to high transport medium volume, allowing the usage of larger volumes of patients’ sample. The bar plot shows Ct values obtained by real time PCR using primer sets N1 and N2 targeting the N gene of SARS-CoV-2 virus. The plot shows two independent experiments, each run in three technical replicates. Primers targeting the human RNase P gene, which amplifies genomic material from human cells were used as positive controls. (N1, N2, and RNase P primers, 2019-nCoV RUO Kit: Integrated DNA Technologies, Cat.N. 10006713). Nasopharyngeal samples from COVID-19 negative donors were spiked in with heat inactivated SARS-CoV-2 viral particles (SeraCare, Cat. N.0505-0126, Lot.N.10481654; or ATCC VR-1986HK) at a concentration of 2.5 cp/µL. Either 5 µL (25%) or 9 µL (45%) of this solution were mixed with 6 µL of AIC mix, heated for 5 minutes at 70°C, and subsequently combined with the RT-PCR master mix (Cytiva One-Step RT-PCR mix).
One of the challenges encountered by public health organizations and diagnostics labs during the current COVID-19 pandemic is the shortage of transport media for sample collection. This has led to inconsistencies associated to COVID-19 sample collection for NAAT. The Center for Disease Control (CDC) recommends alternative collection media that can be prepared locally, including PBS, saline (0.9% NaCl), or the more widely used standard universal viral transport media suitable for cell culture (7,8). The performance of the direct RT-PCR chemistry to detect viral particles in patients’ samples collected in either collection system was tested. Figure 4 shows that the Cytiva direct RT-PCR chemistry allows the detection of viral load at a wide dynamic range irrespective of the transport media used.
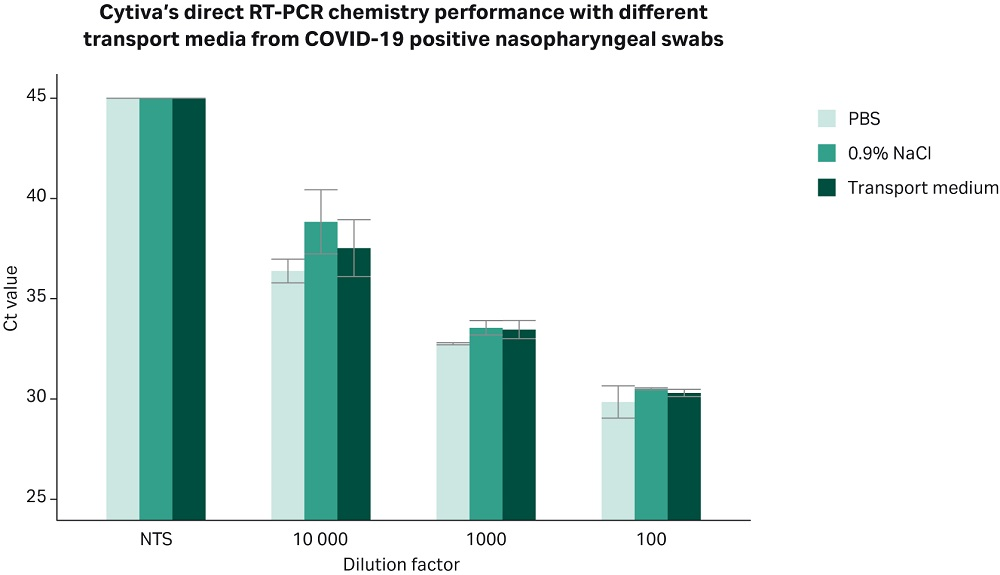
Fig 4. The Cytiva direct RT-PCR chemistry allows robust and sensitive amplification of viral nucleic acids directly from unpurified nasopharyngeal samples contained in a wide range of transport media. Bar plot shows Ct values obtained by real time PCR using primer set N1 targeting the N gene of the SARS-CoV-2 virus. In brief, a pool of COVID-19 negative nasopharyngeal swabs was spiked in with serial dilutions of a pool of nasopharyngeal swabs previously confirmed as positive for COVID-19 by The Clinical Facility Detection at the Kepler Universitätsklinikum, Austria. The samples were contained in either PBS, saline, or commercial transport medium systems. The transport medium system used in this experiment was prepared according to the CDC’s guidelines described in reference (7). All samples were treated as described in previous experiments. In summary, 5 µL of sample were mixed with 6 µL of AIC reagent, heated for 5 minutes at 70 °C, and subsequently combined with primers (N1 primers, 2019-nCoV RUO Kit: Integrated DNA Technologies, Cat.N. 10006713), and Cytiva AIC Mix. Due to limited access to patients’ samples, the plot shows the results from one experiment carried out in triplicate. Bars denote standard deviation of the measurement. X-axis shows the -log10 of the serial dilutions used in the experiment.
The utilization of an extraction-free chemistry for pathogen detection from unpurified samples is a challenge that goes beyond COVID-19 testing. In a proof of principle experiment, we spiked in nasopharyngeal samples from “healthy” donors with synthetic rEbola viral particles (AccuPlex rEbola reference material, SeraCare, Cat.N. 0505-0001, 25 cp/µL of sample). Although this set-up does not represent a bona fide model for the Ebola virus detection, the results show that under the challenging conditions defined by the nasopharyngeal sample, the Cytiva direct RT-PCR chemistry allows for consistent and reliable detection of rEbola viral particles (Fig 5). Further work is being carried out to confirm these results, and to broaden the spectrum of sample types for both clinical and metagenomics applications. This opens an opportunity for assay developers interested in developing an extraction-free NAAT of other organisms and sample types not highlighted in this work.
Fig 5. Direct RT-PCR chemistry from Cytiva allows detection of Ebola synthetic viral particles. Human nasopharyngeal swabs collected in transport media (Nasopharingeal swabs from patients tested negative for COVID-19, Tissue Solutions, FM Reference: QQPE7) were spiked in with 25 cp/µL of heat inactivated rEbola viral particles (SeraCare, AccuPlex™ rEbola GP/NP Reference Material, Item No. 0505-0001). 5 µL of sample were mixed with 6 µL of Cytiva AIC reagent, heated for 5 minutes at 70°C, and subsequently combined with primers (GP gene: primers for hydrolysis probe-based assay were designed based on reference (9), and IDT Primes: RNase P, 2019-nCoV RUO Kit: Integrated DNA Technologies, Cat.N. 10006713), and RT-PCR master mix. The reverse transcription and amplification reactions were carried out in one step, following the kit manufacturer’s recommendations (Cytiva One-Step RT-PCR mix). Bar plot shows Ct values obtained by real-time PCR using primer set GP targeting the GP gene of the Zaire Ebola virus. The plot shows the results combined from two independent experiments ± SD, with three technical replicates run each time. Primers targeting the human RNase P gene, which amplifies genomic material from human cells, were used as positive controls.
Conclusions
The Cytiva direct RT-PCR chemistry, which is a combination of a proprietary AIC and RT-PCR master mix, provides for an extraction-free nucleic acid amplification test (NAAT). This provides a fast, accurate and sensitive detection of viral RNA directly from unpurified nasopharyngeal patient samples, reducing the cost of the assay. More importantly, the time required to deliver result is shortened. Furthermore, the chemistry is versatile and robust: it increases RNA stability in the sample, improves the tolerance to viral transport media, and broadens the spectrum of viral nucleic acids targets and pathogens that can be detected directly from unpurified samples. Using the Cytiva direct RT-PCR chemistry, we were able to detect SARS-CoV-2 viral particles down to approximately 10 copies per reaction. We are carrying out further studies to assess different sample types and pathogens.
Direct RT-PCR chemistry offers a convenient alternative to nucleic acid test (NAT) protocols affected by bottlenecks from shortages of nucleic acid isolation kits and consumables. This approach benefits testing labs looking to streamline their NAT workflow, accelerate high throughput testing, and reduce chances of cross contamination and the associated equipment costs. Until now, the diagnosis of genetic disorders and infectious diseases has required costly and time-consuming procedures. We believe that this chemistry, when used as a diagnostic tool for screening purposes, can help cut down turnaround times to patient diagnosis, supplemented by retesting with an RNA extraction kit of only those patient samples with high suspicion of infection which gave an initial negative result.
For further information on Cytiva direct RT-PCR chemistry, please contact our custom services team, submit an inquiry, or speak to your modality specialist.
Cytiva Direct RT-qPCR Kits are currently available for research use only (RUO).
- Zhang et al. Journal of Medical Virology, 2020: Recent advances in the detection of respiratory virus infection in humans.
- Tahamtan & ARdebili. Expert Review of Molecular Diagnostics, 2020: Real-time RT-PCR in COVID-19 detection: issues affecting the results.
- https://international.neb.com/-/media/nebus/files/application-notes/sars_cov2_detection_an.pdf?rev=6c002a18330747de8a1553bd78f1ea17
- https://www.takarabio.com/products/covid-19-research/viral-detection-with-qpcr/direct-viral-detection-via-qpcr
- He et al. Nature Medicine, 2020. Temporal dynamics in viral shedding and transmissibility of COVID-19.
- Holshue et al. New England Journal of Medicine, 2020. First Case of 2019 Novel Coronavirus in the United States.
- https://www.cdc.gov/coronavirus/2019-ncov/downloads/Viral-Transport-Medium.pdf
- https://www.cdc.gov/coronavirus/2019-ncov/lab/faqs.html
- Pinsky et al. PLOS One, 2015. Analytical Performance Characteristics of the Cepheid GeneXpert Ebola Assay for the Detection of Ebola Virus.