Today tens of thousands of individual cells from a single tissue sample or patient can be analyzed, giving researchers an unprecedented opportunity to understand individual cell populations and their behavior in diseased tissue. This is critical for analyzing complex, heterogeneous biological systems from cancer to forensics and analyzing individual cells to aid in monitoring disease or to guide therapy strategies. One of the first and most critical steps in an scRNA-seq protocol is the dissociation of tissues to yield fully dissociated but intact and viable cells.
Manual tissue dissociation is long, laborious and affects cell viability. It is vital to choose equipment to speed up the single-cell workflow yet at the same time be gentle on the cells and preserve the original cell state as much as possible. Many mechanical dissociation platforms on the market focus on warm dissociation. Preliminary research has shown that cold-acting enzymatic dissociation reduces cell stress (1) therefore minimizing cell death and by consequence improving the transcriptional profile of the single cells.
The Cold dissociation enzyme mix – kidney (the Cytiva kit) comprises of two enzymes and a buffer. When used with the VIA Extractor™ tissue disaggregator, the Cytiva kit improves cellular viability, reduces cellular aggregation, maintains RNA stability and reduces cell stress in comparison to warm enzymes. The VIA Extractor tissue disaggregator can sustain a cold temperature in a controlled manner making it ideal for dissociation at cooler temperatures. The Cytiva kit has been developed for use with the VIA Extractor™ tissue disaggregator at cold temperatures.
The following data was generated to demonstrate the effectiveness of the Cytiva kit for disaggregation of mouse kidney into a viable single-cell suspension that is suitable for use in single-cell RNA sequencing. The Cytiva kit dissociation on the VIA Extractor™ tissue disaggregator was compared to warm enzyme dissociation using Miltenyi Biotec’s Multi tissue dissociation kit 2 (the Miltenyi kit).
Comparative performance of the Cold dissociation enzyme mix – kidney to Miltenyi Biotec’s Multi tissue dissociation kit 2
Mouse kidneys were obtained from 6 female Crl:CD1 (ICR) mice and washed in ice cold Dulbecco’s phosphate buffered saline (DPBS). Any connective tissue was removed. The mouse kidneys were used as paired samples: one for dissociation with Cytiva kit and the other for dissociation with Multi tissue dissociation kit 2. Both pairs were dissociated on the VIA Extractor tissue disaggregator. Equal amounts of tissue and enzyme mix were added to each experimental replicate (Fig 1 and Table 1).
| User | Mouse | Enzyme temperature | Enzyme kit used | Kidney weight | Volume of enzyme mix |
|---|---|---|---|---|---|
| 1 | 1 | Cold | Cytiva Cold dissociation enzyme kit - kidney | 299 mg | 5 mL |
| 1 | 2 | Cold | Cytiva Cold dissociation enzyme kit - kidney | 245 mg | 5 mL |
| 1 | 3 | Cold | Cytiva Cold dissociation enzyme kit - kidney | 267 mg | 5 mL |
| 2 | 1 | Cold | Cytiva Cold dissociation enzyme kit - kidney | 226 mg | 5 mL |
| 2 | 2 | Cold | Cytiva Cold dissociation enzyme kit - kidney | 247 mg | 5 mL |
| 2 | 3 | Cold | Cytiva Cold dissociation enzyme kit - kidney | 239 mg | 5 mL |
| 1 | 4 | Warm | Miltenyi Biotec Multi Tissue Dissociation Kit 2 | 284 mg | 5 mL |
| 1 | 5 | Warm | Miltenyi Biotec Multi Tissue Dissociation Kit 2 | 266 mg | 5 mL |
| 1 | 6 | Warm | Miltenyi Biotec Multi Tissue Dissociation Kit 2 | 252 mg | 5 mL |
| 2 | 4 | Warm | Miltenyi Biotec Multi Tissue Dissociation Kit 2 | 200 mg | 5 mL |
| 2 | 5 | Warm | Miltenyi Biotec Multi Tissue Dissociation Kit 2 | 256 mg | 5 mL |
| 2 | 6 | Warm | Miltenyi Biotec Multi Tissue Dissociation Kit 2 | 248 mg | 5 mL |
Table 1. Tissue weights and enzyme mix volumes for each sample dissociated with cold and warm enzyme mixes. Note that two experiments were performed with two separate users on two separate occasions.
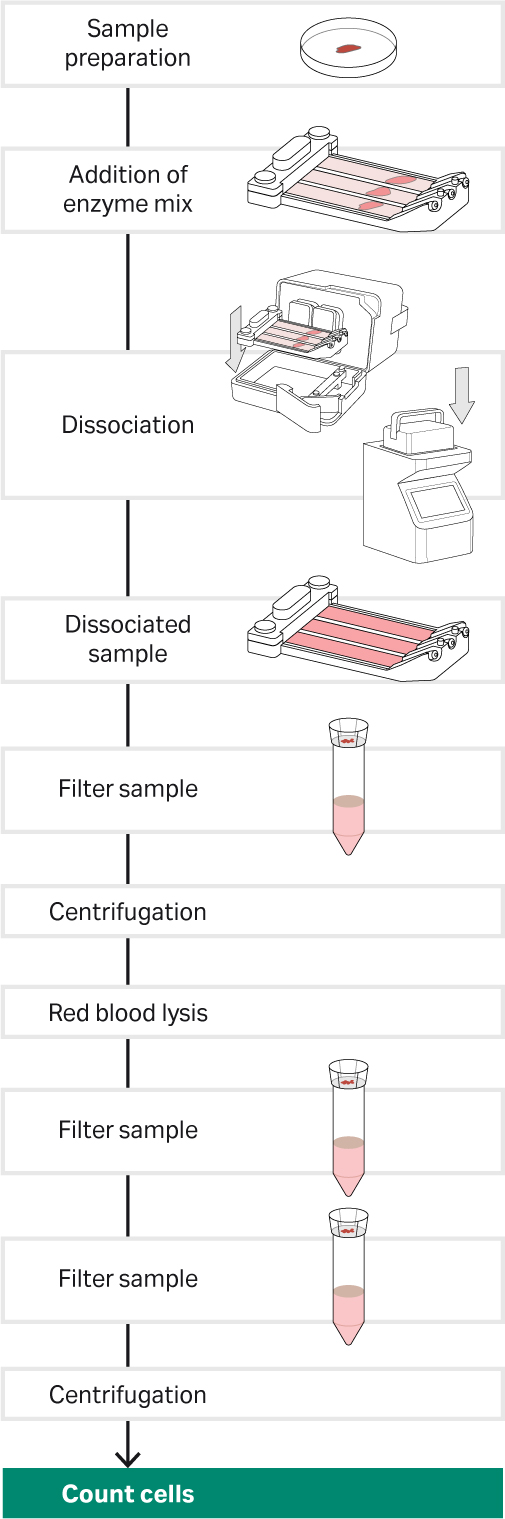
Fig 2.Tissue dissociation workflow for the Cytiva kit and Miltenyi kit the VIA Extractor tissue disaggregator. Note the procedure had 3 sample filter steps. The first with 100 µm cell strainer, the second with 70 µm cell strainer and the third with 40 µm cell strainer.
| Cytiva Cold dissociation enzyme kit – kidney |
Miltenyi Biotec Multi Tissue Dissociation Kit 2 | |
|---|---|---|
| Program speed | 200 rpm | 200 rpm |
| Program time | 15 mins | 15 mins |
| Program temperature | 2°C | 36°C |
Table 2. Temperature, speed and time conditions for Cytiva kit and warm enzyme on the VIA Extractor tissue disaggregator. Note the program temperature is set at 2°C to allow the VIA Extractor tissue disaggregator to cool to 4˚°C .
For warm tissue dissociation with The Miltenyi kit, the enzyme volumes in Miltenyi’s Biotec’s “Dissociation of mouse kidney using the Multi Tissue Dissociation Kit 2” protocol were followed. For cold dissociation using Cytiva kit on the VIA Extractor tissue disaggregator, the Cytiva kit procedure was followed.
All cell suspensions were passed through 100 µm cell strainers and subjected to red blood cell lysis. Following red blood cell lysis, cells were resuspended in DPBS supplemented with 0.4% (w/v) bovine serum albumin (BSA) and 0.1 mM ethylenediaminetetraacetic acid (EDTA) and filtered through 70 µm then 40 µm cell strainers. Cells were counted using a Nucleocounter NC-200™ and VIA2-Cassettes™ (ChemoMetec). All t-tests were performed in JMP® Statistical Discovery™ software by SAS.
Two experiments were performed with different users on separate occasions. For the first occasion, three paired samples were subjected to single-cell RNA sequencing to compare transcripts and cell types between cold and warm enzymes. Cells from the second occasion were not sequenced.
Results for comparative performance of the Cold dissociation enzyme mix –kidney kit to Miltenyi Biotec’s Multi tissue dissociation kit 2
Use of the Cytiva kit on VIA Extractor tissue disaggregator results in higher viability and fewer aggregated cells than when using the Miltenyi kit (Fig 2, A and B). Probability values (p values) remained statistically significant when adjusted for multiple testing using Bonferroni correction (3 tests performed). For single cell sequencing, to acquire the best quality data that represents the true nature of the cell, it is important to have high viability cells and a low rate of cell aggregates to increase capture of single cells rather than multiplets. These data show that the Cold dissociation enzyme mix - kidney promotes cell viability and reduces the number of cells stuck together.
It is worth noting that the yield of cells when using The Miltenyi kit is higher than when using the Cytiva kit. Nonetheless, the lower yield of cells has no impact on the cell types detected by single-cell RNA sequencing.
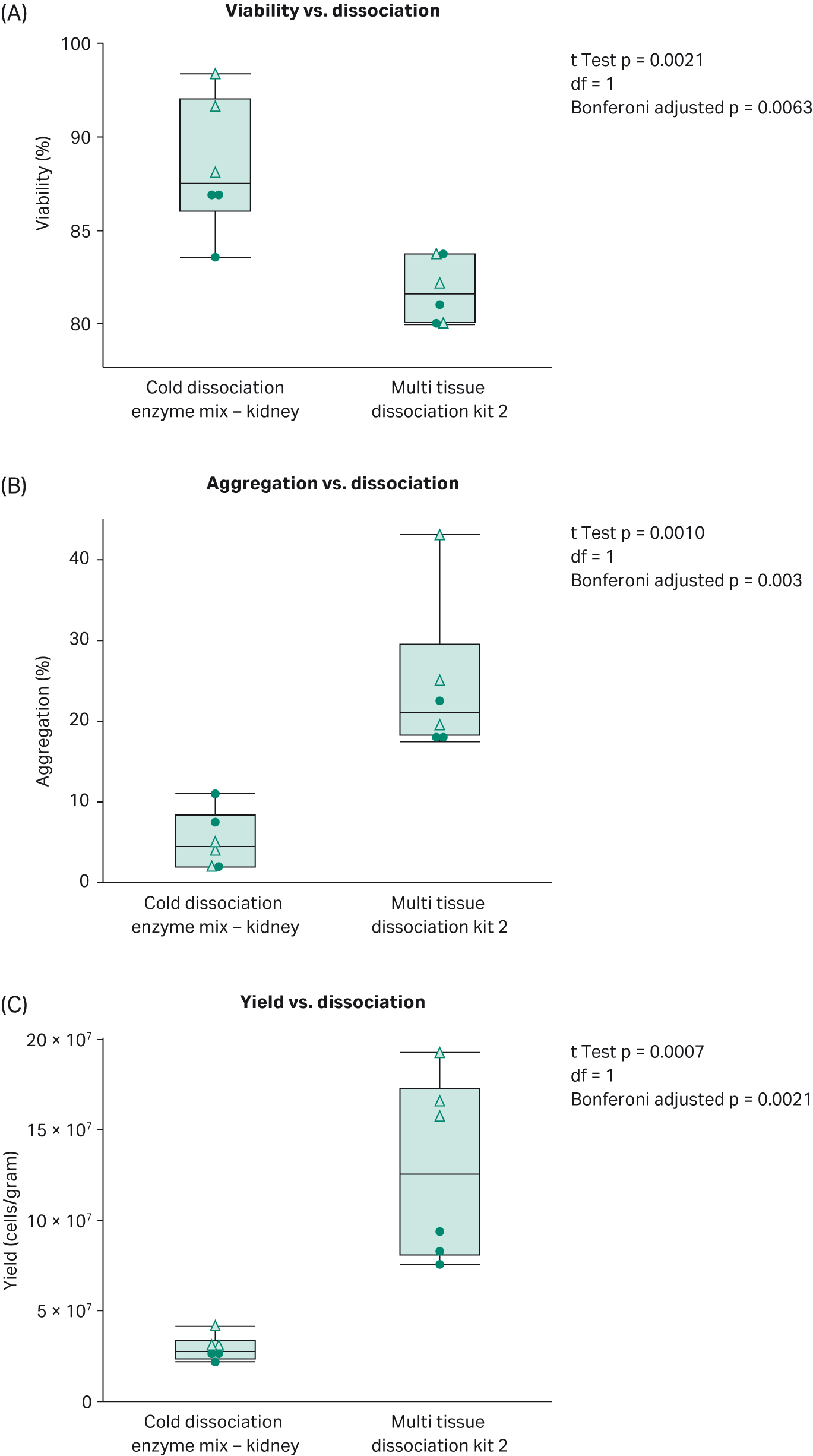
Fig 3.Comparison of (A) percentage viability, (B) percentage aggregation and (C) cell yield between dissociation on VIA Extractor tissue disaggregator when using the Cytiva kit and Multi tissue dissociation kit 2 . Dots represent experiment by user 1 and triangles experiment by user 2. Use of the Cytiva kit, results in statistically significantly lower aggregation, higher viability but lower cell yield compared with The Miltenyi kit.
Summary of method for single-cell RNA sequencing
For single-cell sequencing, cells from samples were quantified to ascertain mean count. The manufacturer’s instructions for 10x Genomics Chromium Next GEM Single Cell 3’ dual index kit v3.1 were followed with an aim to sequence 1000 cells for each sample. A 10x Genomics Chromium Controller was used to capture the cells into gel beads in emulsion (GEMs). Libraries were sequenced on a NextSeq 550 Base (Illumina Inc.) using NextSeq 550 high output kit v2.5 (Illumina Inc.). Two sequencing runs were performed to achieve depth. The scRNA matrix data were analyzed using UMAP (2) in Seurat (3). Each sample was analyzed individually and filtered to remove duplicates and include cells with feature RNA between 200 and 4000 and keep all cells with a mitochondrial gene expression percentage less than 50%. Once all samples were filtered and clustered, the data from each sample were combined into a single dataset to allow comparison of the Cytiva kit with the Multi tissue dissociation kit 2 data using Seurat (3) and UMAP (2). Cell types representing each cluster were identified using Seurat and marker genes identified by He et al. (4) and Chung et al. (5). Genes lists from cell clusters with differential gene expression profiles were further analyzed using gene ontology software package PANTHER (6). All t-tests were performed in JMP® Statistical Discovery™ software by SAS.
Results for single-cell RNA sequencing
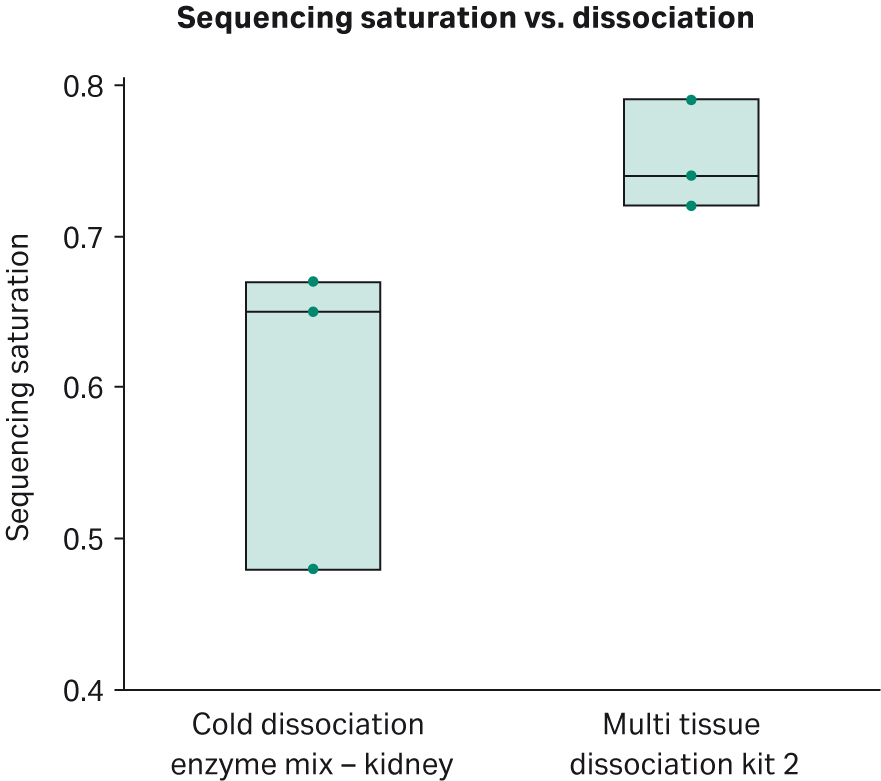
Samples from the Cold dissociation enzyme mix - kidney and The Miltenyi kit performed the same for 10X single-cell RNA GEM preparation and library preparation. There were no differences in cDNA concentration or cDNA quality observed in bioanalyzer traces. Also, there were no differences in library concentration or quality. For single cell sequencing metrics, there were no statistical differences in passing cells (average 2320; Fig 3), mitochondrial gene expression, threshold for passing cells (average 797 per cell) and mean reads per cell (average of 26828 read per cell). For passing cells, sample 3 for the Cold dissociation enzyme mix - kidney was an outlier; nonetheless, differences in passing cells were none significant (t-test = -0.68 , degrees of freedom=4, p=0.536). Although there were no differences in number of passing cells and other single-cell sequencing metrics, for saturation, there was a statistically significant increase in sequencing saturation for The Miltenyi kit compared with the Cold dissociation enzyme mix - kidney (t Test = 2.35, degrees of freedom= 4, p=0.0392; Fig 4). Although the p value was not statistically significant when corrected for multiple testing, these data highlight a trend for increased sequencing saturation from warm dissociation. Sequencing saturation is an indicator of cellular content of RNA and library complexity. For example, if a score of 50% saturation is given, potentially only half the transcript information is captured for each cell. Cells with high transcriptional activity and higher RNA complexity require increased depth of sequencing to capture the total transcriptomic information. Both dissociation methods were sequenced at the same time with the same methods and conditions and there were no differences in number of cells captured, in cDNA concentrations, or cDNA profiles from single-cell library preparation. These data imply that cells from kidneys dissociated with Cytiva’s Cold dissociation enzyme mix - kidney may contain a wider breadth of transcriptomic information.
Our single-cell RNA sequencing data support the notion that the Cold dissociation enzyme mix - kidney promotes RNA stability. Cluster profiles between Cytiva’s Cold dissociation enzyme mix - kidney and The Miltenyi kit are very similar (Fig 5 and Fig 6). All major cell types were detected: immune cells, proximal tubules, mesangial, distal tubules (with alpha and beta intercalated cells), podocytes and glomeruli. Interestingly, for Cytiva’s kit, a high proportion of cells in cluster 21 were detected in comparison with The Miltenyi kit.
Table 3 lists the top ten differentially expressed genes in cluster 21. HBA-A2 , HBB-BT, HBA-A1, MKRN1, NUSAP1, BPGM and HBB-BS are all genes reported to be expressed in granulocytes. Granulocytes, a key component in innate immunity, are first line of defence in an immune response. Granulocytes are defined by their granular appearance and fall into three categories: neutrophils, basophils and eosinophils. Granulocytes do not yield high quantities of RNA yet they contain high concentrations of RNases (8-9) and this is likely to pose a challenge in single-cell RNA sequencing. Performing the dissociation at cold temperatures and carrying out all subsequent cell clean up steps on ice, is likely to reduce RNase activity and preserve the granulocyte.
Three mitochondrial genes were also detected in the top ten differentially expressed genes in cluster 21 (Table 3). UCP2, GPX4 AND ATP5B proteins are involved in oxidative stress regulation and in Cytiva’s Cold enzyme dissociation mix – kidney had negative fold change levels implying that the levels of these transcripts was lower in cluster 21 compared to other clusters.
Transcription from cellular stress response genes has been reported to be higher in cells isolated from tissue dissociated in warm conditions compared to cold temperatures (O’Flanagan et al 2010). To determine if there were any differences in cell stress gene expression between Cytiva’s Cold dissociation enzyme mix and Miltenyi’s Multi tissue dissociation kit 2, dot plots were created using Seurat to display the proportion of the cells in each cluster expressing the genes described in O’Flanagan’s research study (Figure 7 A). Clusters 10, 13, 15 and 19, displayed a high proportion of cells expressing these genes and correlate to monocytes, T cells and podocytes. Further differential expression analysis support that key cell stress markers are expressed at higher levels in Miltenyi’s Multi tissue dissociation kit 2 compared to Cytiva’s Cold dissociation enzyme mix (Figure 7B). These data emphasise that Cytiva’s Cold dissociation enzyme mix reduces cellular stress response during dissociation. ID1 gene was expressed at a higher proportion and level in podocytes for cells dissociated with Cytiva’s cold dissociation mix compared to Miltenyi’s warm Multi tissue dissociation kit 2. It is unclear what role ID1 plays in podocytes. ID1 regulates cellular differentiation and the cell cycle. Overexpression of ID1 has been reported to delay the onset of endothelial senescence (10). It is plausible the ID1 exerts a positive effect on podocytes by promoting proliferation and delaying senescence. Further studies are required to fully understand the role of ID1 in the kidney.
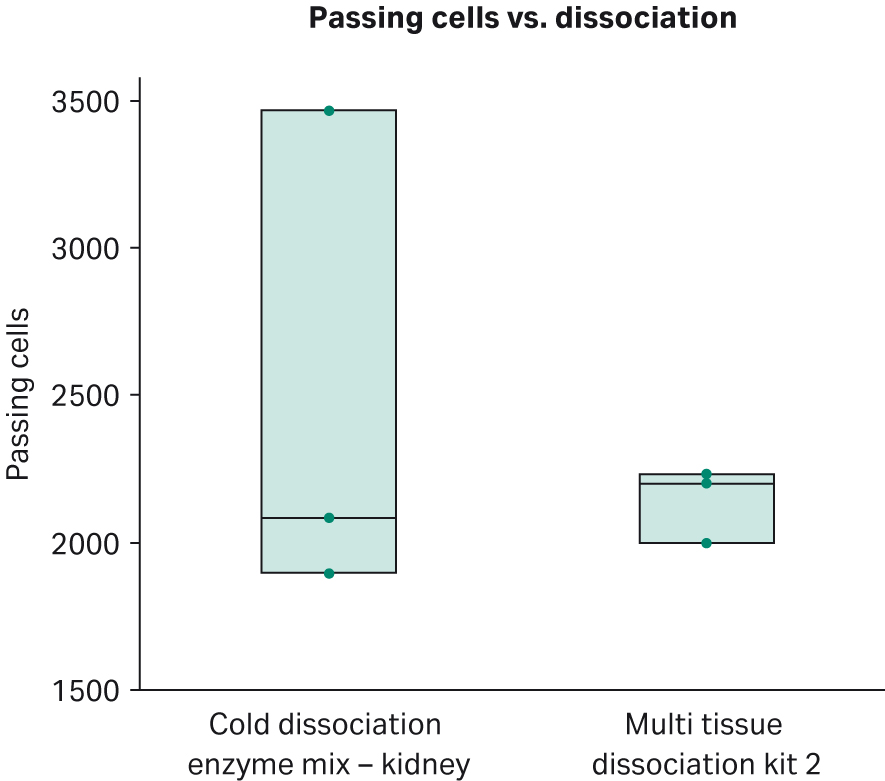
Fig 4.Number of passing cells for mouse kidney dissociated with the Cold dissociation enzyme mix - kidney and Miltenyi Biotec’s Multi Tissue Dissociation Kit 2.The Cold dissociation enzyme mix - kidney has one outlier where approximately 1500 more cells were loaded. There is no statistically significant difference in passing cells between the two groups t-test = -0.68, degrees of freedom=4, p=0.536.
Fig 5.Sequencing saturation comparison between tissue dissociated with the Cold dissociation enzyme mix - kidney and Miltenyi Biotec’s Multi Tissue Dissociation Kit 2. The Miltenyi kit shows a higher sequencing saturation than the Cold dissociation enzyme mix - kidney (t-test = 2.35, degrees of freedom= 4, p=0.0392; p = 0.314 when adjusted for multiple testing using Bonferroni with 8 tests). Note that this result remains significant when sample 3 from the Cold dissociation enzyme mix - kidney is removed from analysis (t-test = 3.23, degrees of freedom= 3, p=0.0483; p=0.386 when adjusted for multiple testing using Bonferroni with 8 tests).
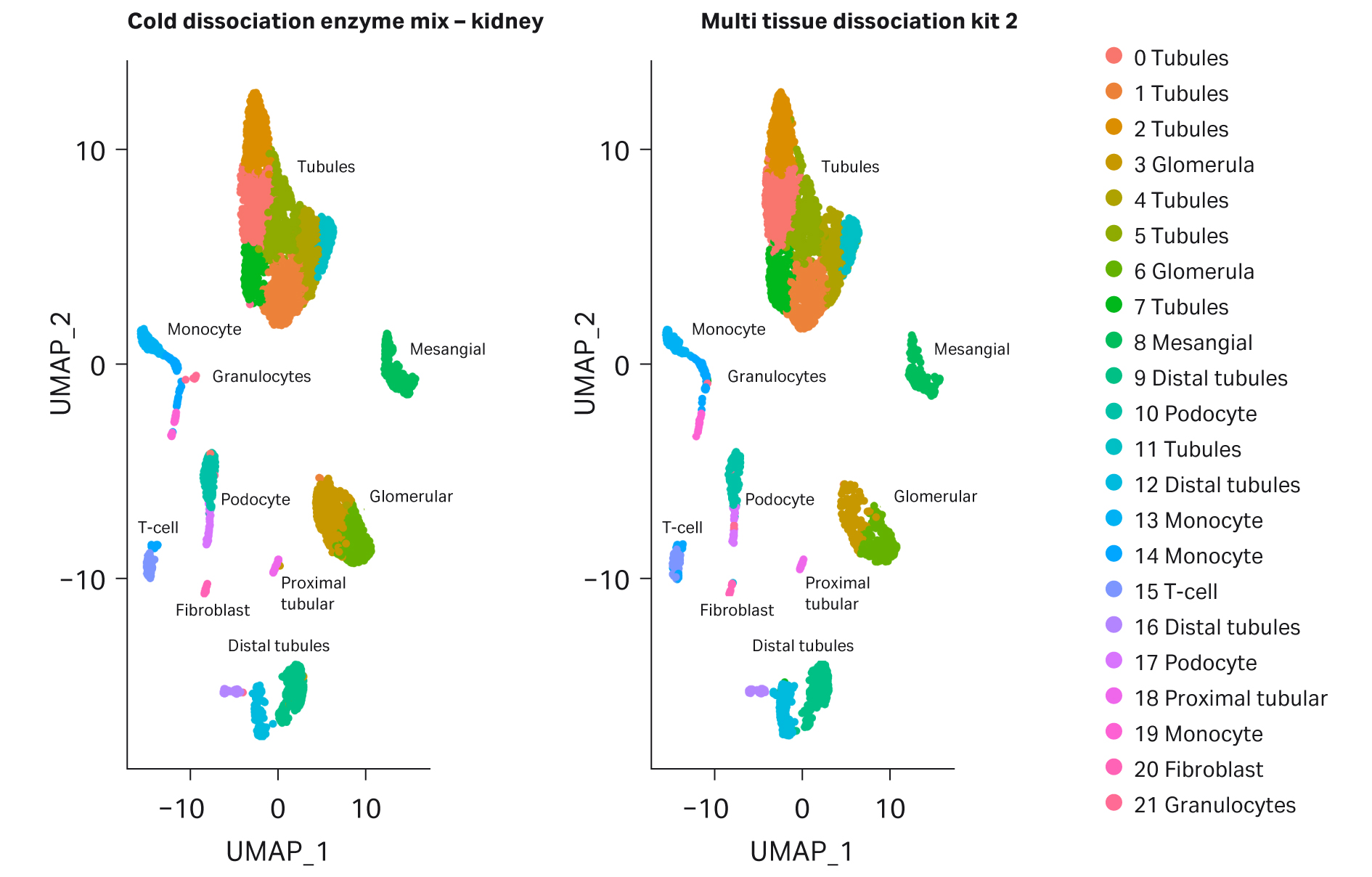
Fig 6.UMAP plots comparing kidney cells dissociated with Cytiva's Cold dissociation enzyme mix – kidney and Miltenyi Biotec's Multi Tissue Dissociation Kit 2
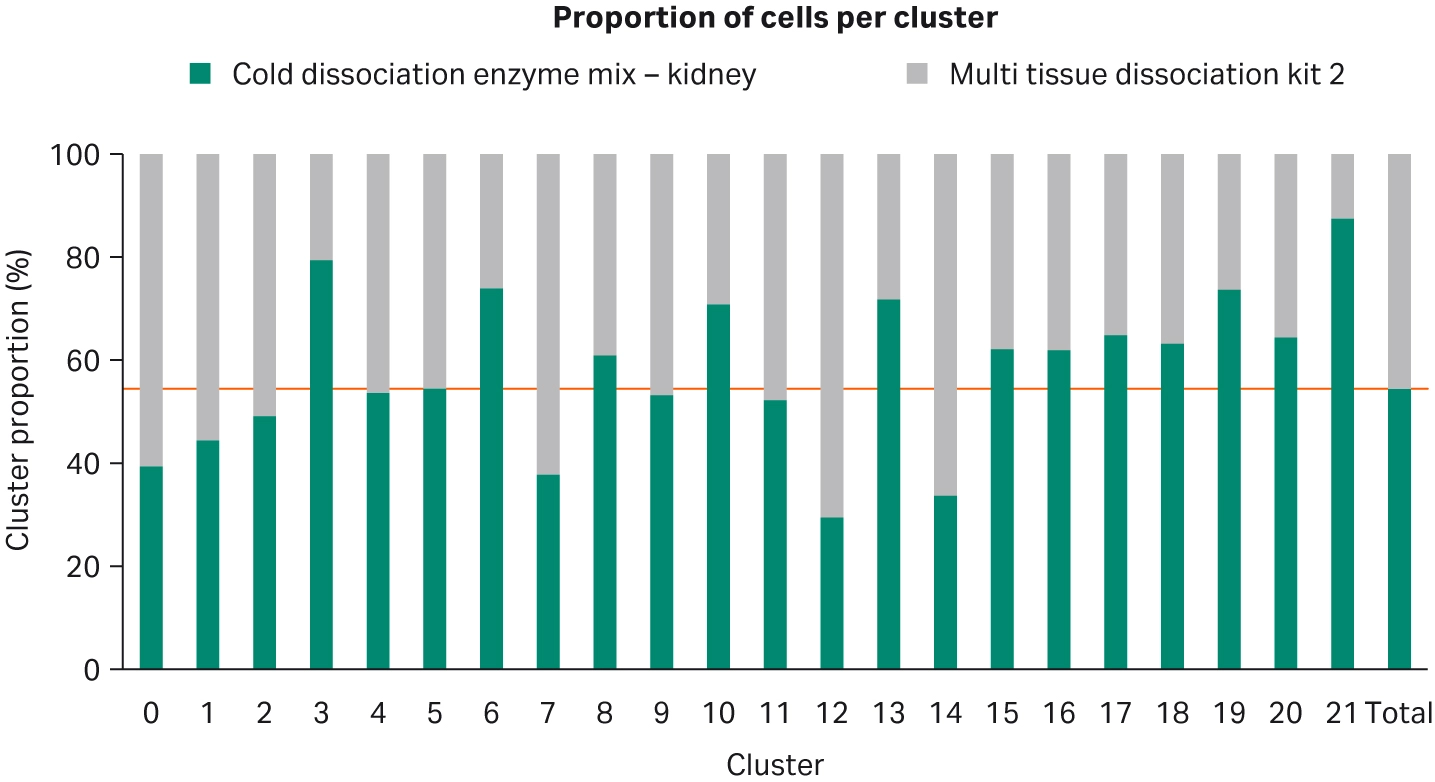
Fig 7.Bar chart showing proportion of cells dissociated with Cytiva's Cold dissociation enzyme mix - kidney and Miltenyi Biotec's Multi Tissue Dissociation Kit 2 for each cluster. There is a higher proportion of cluster 21 cells for dissociation carried out with Cytiva’s Cold dissociation enzyme mix - kidney.
| Transcript ID | Gene ID | Gene name | Miltenyi Multi tissue dissociation kit 2 Log fold change | Cytiva Cold dissociation enzyme mix Log fold change | Cluster minimum p value | cell type | role |
|---|---|---|---|---|---|---|---|
| ENSMUSG00000069917 | HBA-A2 |
Alpha globin A2 |
1.78 |
5.03 | 0 | granulocytes |
unknown function in granulocytes |
| ENSMUSG00000069917 |
HBB-BT |
Alpha globin A2 |
2.89 |
8.51 | 2.57x10-108 |
granulocytes |
unknown function in granulocytes |
| ENSMUSG00000069919 |
HBA-A1 |
Alpha Globin A1 |
4.62 | 8.48 | 2.22x10-45 |
granulocytes |
unknown function in granulocytes |
| ENSMUSG00000069919 |
MKRN1 |
Makorin ring finger protein 1 |
0.32 | 3.55 | 2.41x10-38 |
granulocytes |
controls cell cycle arrest and apoptosis |
| ENSMUSG00000027306 |
NUSAP1 |
Nucleolar and spindle associated protein 1 |
2.02 | 0.41 | 1.12x10-36 |
granulocytes |
Cell Cycle |
| ENSMUSG00000038871 |
BPGM |
Bisphosphoglycerate mutase |
-0.43 | 3.36 | 5.83x10-34 |
granulocytes |
unknown function in granulocytes |
| ENSMUSG00000052305 |
HBB-BS |
Bisphosphoglycerate mutase |
5.68 | 8.68 | 1.50x10-28 |
granulocytes |
unknown function in granulocytes |
| ENSMUSG00000033685 |
UCP2 |
Uncoupling protein 2 |
1.33 | 1.07 | 1.07x10-19 |
mitochonrial |
Oxidative phosphorlyation |
| ENSMUSG00000075706 |
GPX4 |
Glutathione peroxidase 4 |
0.43 | -3.07 | 2.88x10-17 |
mitochonrial |
Oxidative stress protection |
Table 3. Top ten differentially expressed genes defining cluster 21
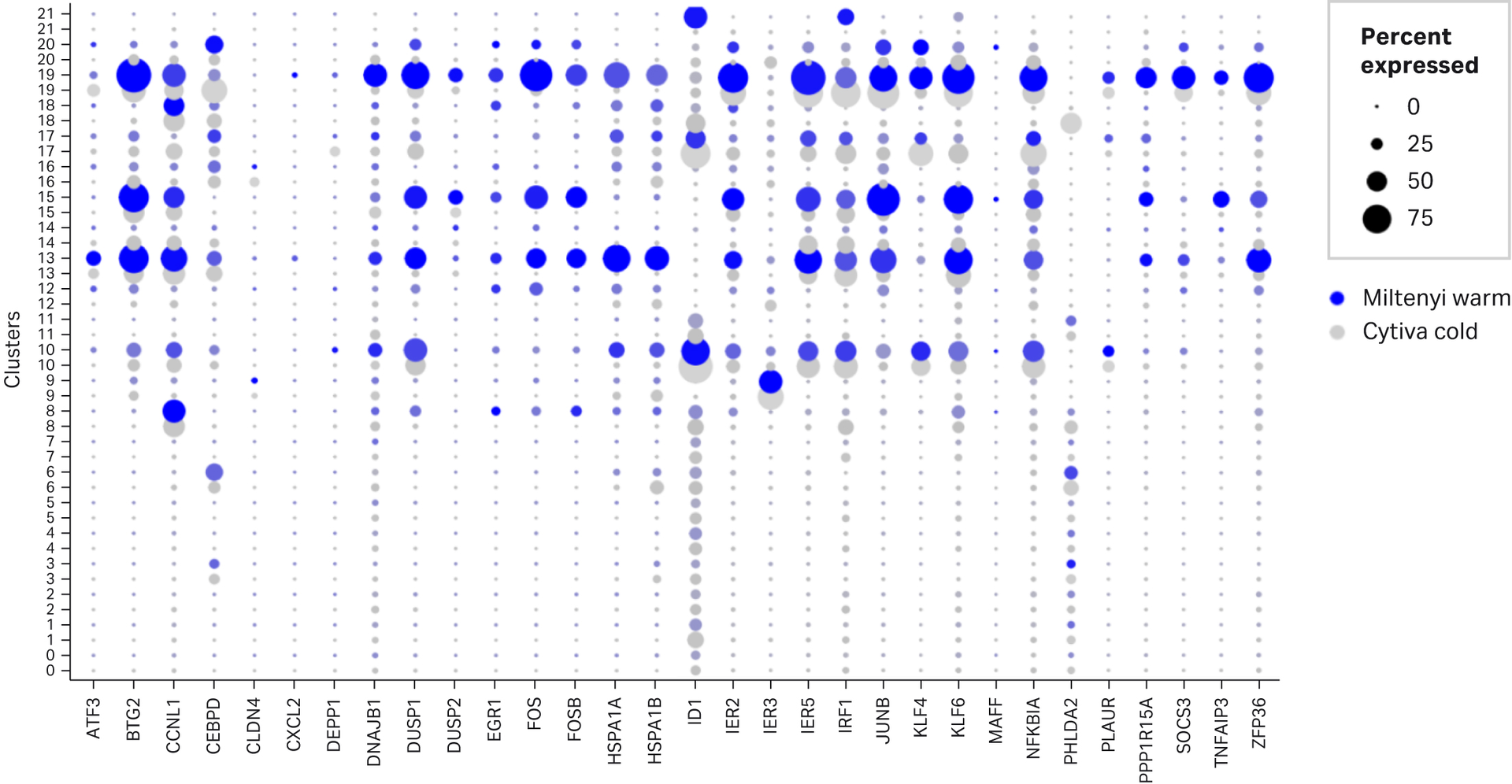
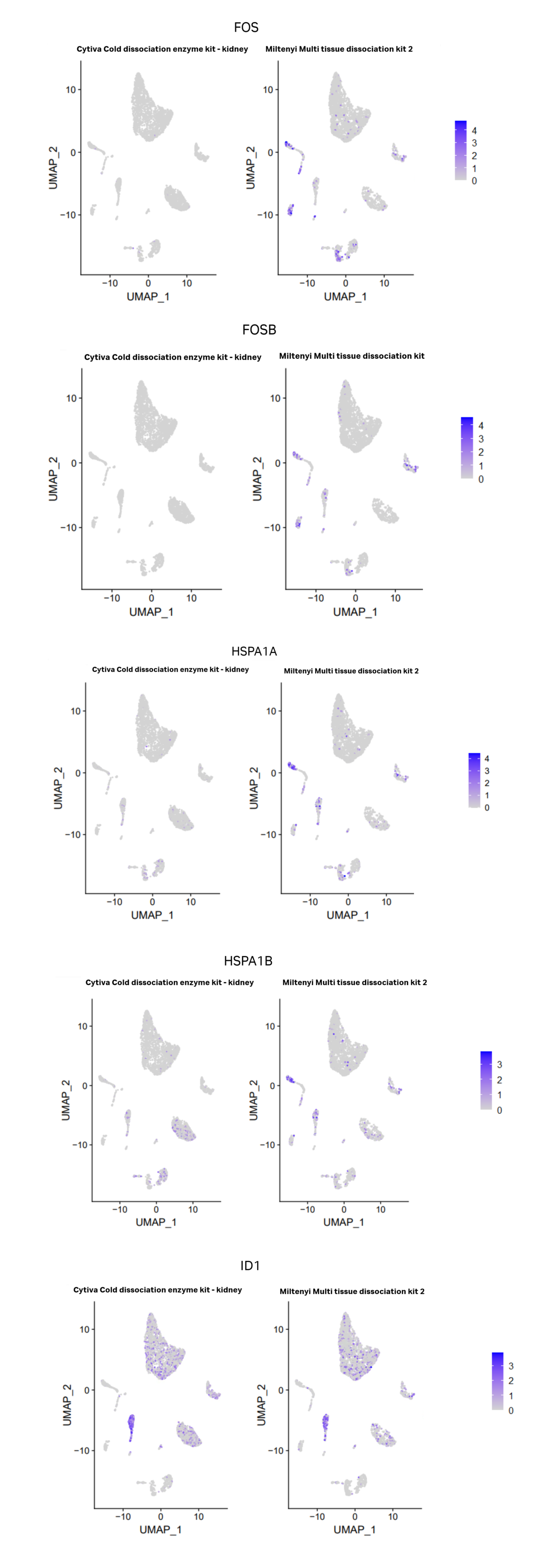
Fig 8. Differential stress marker gene expression between Miltenyi’s multi tissue dissociation kit 2 and Cytiva’s Cold dissociation enzyme mix - kidney. A. Dot plots showing percent of cells in each cluster expressing “stress” gene's listed in O’Flanagan et al (2019) study. Blue dots represent Miltenyi’s multi tissue dissociation kit 2 and grey represent Cytiva’s Cold dissociation enzyme kit. With the exception of ID1, there is a higher percentage of cells expressing the majority of stress genes in Miltenyi’s warm enzymes compares to Cytiva’s cold. B. Feature plots displaying differences in expression of 4 key stress response genes FOS, FOSB, HSPA1A, HSPA1B between Miltenyi’s multi tissue dissociation kit 2 and Cytiva’s Cold dissociation enzyme mix - kidney. A feature plot for ID1 is included due to increased percentage of cells expressed in dot plot. The scales show maximum expression value for each feature. With the exception of ID1, Miltenyi’s warm enzymes show increased expression of FOS, FOSB, HSPA1A and HSPA1B. ID1 appears to have increased expression in podocytes for Cytiva’s cold enzymes compared to Miltenyi’s warm enzymes.
Conclusion
There is growing evidence to support cold enzymatic dissociation is beneficial to preserve cell state and reduce cell stress during the dissociation process. Here we present data that demonstrates Cytiva’s Cold dissociation enzyme mix - kidney has multiple benefits for the single-cell researcher: Cytiva’s Cold dissociation enzyme mix - kidney reduces clumping of cells and improves cell viability. A lower yield given by Cytiva’s Cold dissociation enzyme mix - kidney has no impact on the cell types detected and the same cell types were detected for both enzymatic dissociation methods. Cytiva’s Cold dissociation enzyme mix - kidney may also reduce RNA degradation, preserve the transcriptome and allow detection of cells difficult to sequence due to high RNase content.
Ordering information
| Product |
Description |
Code |
|---|---|---|
| Cold Dissociation Enzyme Mix – Kidney | An enzymatic kit for kidney tissue dissociation at 4⁰C, optimized for semi-automated disaggregation in single-cell workflows. |
29733434 1 pack x 20 reactions |
| Omics Bundle | VIA Extractor™* tissue disaggregator, VIA Freeze™ Uno controlled-rate freezer and Omics clamp in one convenient package. VIA Freeze™ Uno controlled-rate freezer is a liquid nitrogen-free controlled-rate freezer |
29517120 |
| Omics clamp | Works with Omics pouch providing a secure seal after the sample insertion whilst providing the support to the bag during tissue dissociation in the VIA Extractor™ tissue disaggregator | 29509355 |
| Omics pouch | Single-use, multi-compartment bag, for dissociation of tissue into single-cell suspension. Contains 3 individual compartments and tubing to allow the addition of digestive enzyme solutions. Each compartment can contain up to 1.2 g of sample and 2 to 5 mL of total volume | 29726921 (10 pack) |
| Omics applicator | Syringe mechanism to aid insertion of tissue sample into the Omics pouch for enzymatic and mechanical tissue dissociation using VIA Extractor™ tissue disaggregator |
29509359 |
2. Becht E, McInnes L Healy J, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018 Dec 3. doi: 10.1038/nbt.4314. Online ahead of print.
3. Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell 2021; Jun 24;184(13):3573-3587
4. He B, Chen P, Zambrano S, et al. Single-cell RNA sequencing reveals the mesangial identity and species diversity of glomerular cell transcriptomes Nat Commun 2021; 12, 2141
5. Chung J, Goldstein L, Chen YJ, et al. Single-Cell Transcriptome Profiling of the Kidney Glomerulus Identifies Key Cell Types and Reactions to Injury .J Am Soc Nephrol 2020;Oct;31(10):2341-2354
6. Huaiyu M, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res2019 ;Jan 8;47(D1):D419-D426
7. Yang A, Miller DM. Purification of functional RNA from human granulocytes. J Lab Clin Med. 1985 Jan;105(1):94-8. PMID: 2578534.
8. Rosenberg HF, Dyer KD, Foster PS. Eosinophils: changing perspectives in health and disease. Nat Rev Immunol. 2013 Jan;13(1):9-22. doi: 10.1038/nri3341. Epub 2012 Nov 16. PMID: 23154224; PMCID: PMC4357492.
9. Bechmann VG, Hansen AT , Poulsen LK , Jensen BM. Quantity and Quality of Basophil RNA Depend on the RNA Extraction Technique. Methods Mol Biol. 2020;2163:241-245
10. Tang J, Gordon GM, Nickoloff BJ, Foreman KE. The Helix–Loop–Helix Protein Id-1 Delays Onset of Replicative Senescence in Human Endothelial Cells. Lab Inv. 2002 April; 82(8):1073-1079