By Devina Divekar, Post Doctoral Genomics Research Fellow, Cytiva
and Rodrigo Grandy-Morgan, Senior Development Scientist - Genomics and Cellular Research, Cytiva
Single-cell sequencing is a powerful tool in the study of cellular heterogeneity among individual cells. Advances in single-cell RNA sequencing (scRNA-seq), in particular, have enabled scientists to gain unprecedented insights into transcription profiles at the single-cell level, allowing identification and study of rare cell populations in tissues of interest. Due to the high sensitivity of single-cell analyses, such as scRNA-seq, detailed attention must be put into experimental setup and execution. Careful handling and processing of human or animal tissue sample is critical to minimize any process-induced effects that may skew results. Here, we investigate the ability of the VIA Extractor™ tissue disaggregator (Cytiva)* (Fig.1) to generate suspensions of high-quality single cells from fresh tissue for use in scRNA-seq analysis, and compare performance to the gentleMACS™ dissociator (Miltenyi Biotec).
The VIA Extractor™ tissue disaggregator provides fast, low impact tissue dissociation into single-cell suspensions, (B) The Omics pouch placed into the VIA Extractor™ tissue disaggregator and held in place with the Omics clamp. (C) The VIA Extractor™ tissue disaggregator placed into the top of the VIA Freeze™ Uno controlled-rate freezer.
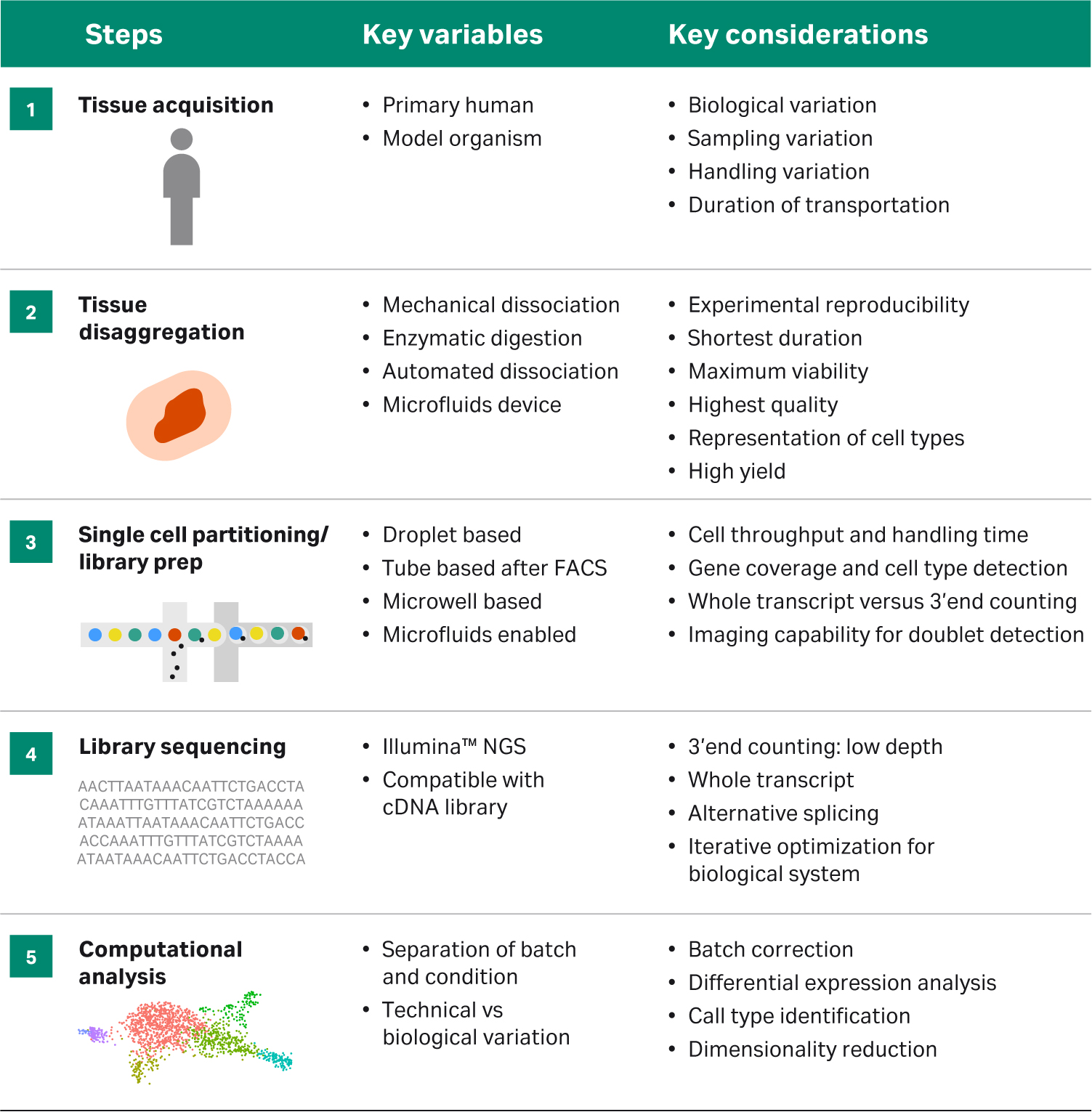
Tissue processing is extremely important in achieving the best results for any downstream single-cell application. In order to obtain a high yield of healthy viable cells, it is important to isolate the tissue of interest while it is as fresh as possible and to process immediately after collection. During processing, cells are continually responding to changes in their environment, that can carry over into transcription profiles expressed by those cells, so minimizing the impact of this processing prior to library preparation is key to ensuring that the best quality data is obtained downstream. A fast, yet gentle tissue dissociation method that is reproducible will help avoid any batch effect or variation due to handling. Table 1 describes the variables and considerations to keep in mind while planning any single-cell sequencing experiment.
Table 1 Overview of stepwise approach to designing single-cell analysis workflow. Adapted from: https://www.frontiersin.org/files/Articles/391125/fcell-06-00108-HTML/image_m/fcell-06-00108-g001.jpg
Murine liver tissue was collected from three freshly dissected 129Sv females. After careful isolation the tissue was weighed and washed with culture media and liver lobes from each mouse were carefully and equally divided between the VIA Extractor™ tissue disaggregator and gentleMACS™ to minimize the sample variability (Table 2). Tissue disaggregation protocols were performed according to manufacturer’s instructions (Table 3).
All mice were chosen as littermates to minimize variability between samples.
Table 2 Tissue weights and the volume of enzyme mix used per sample in both gentleMACS™ and VIA Extractor™ tissue disaggregator.
| Mouse | Liver for gentleMACS™ | Liver for VIA Extractor™ tissue disaggregator | Enzyme volume |
|---|---|---|---|
| 1 | 400 mg | 396 mg | 4 mL gentleMACS™ enzymes |
| 2 | 4 mL gentleMACS™ enzymes | 398 mg | 4 mL gentleMACS™ enzymes |
| 3 | 399.2 mg | 369 mg | 4 mL gentleMACS™ enzymes |
Table 3 Liver tissue was dissociated using the manufactures guidelines defining the parameters such as speed, time and temperature.
| Mouse liver | gentleMACS™ | VIA Extractor™ tissue disaggregator |
|---|---|---|
| Processing speed (RPM) | N/A | 200 |
| Time to complete digestion (minutes) | ~37 secs processing + 30 minute incubation + ~37 secs processing | 10 |
| Processing temperature | 37°C | 37°C |
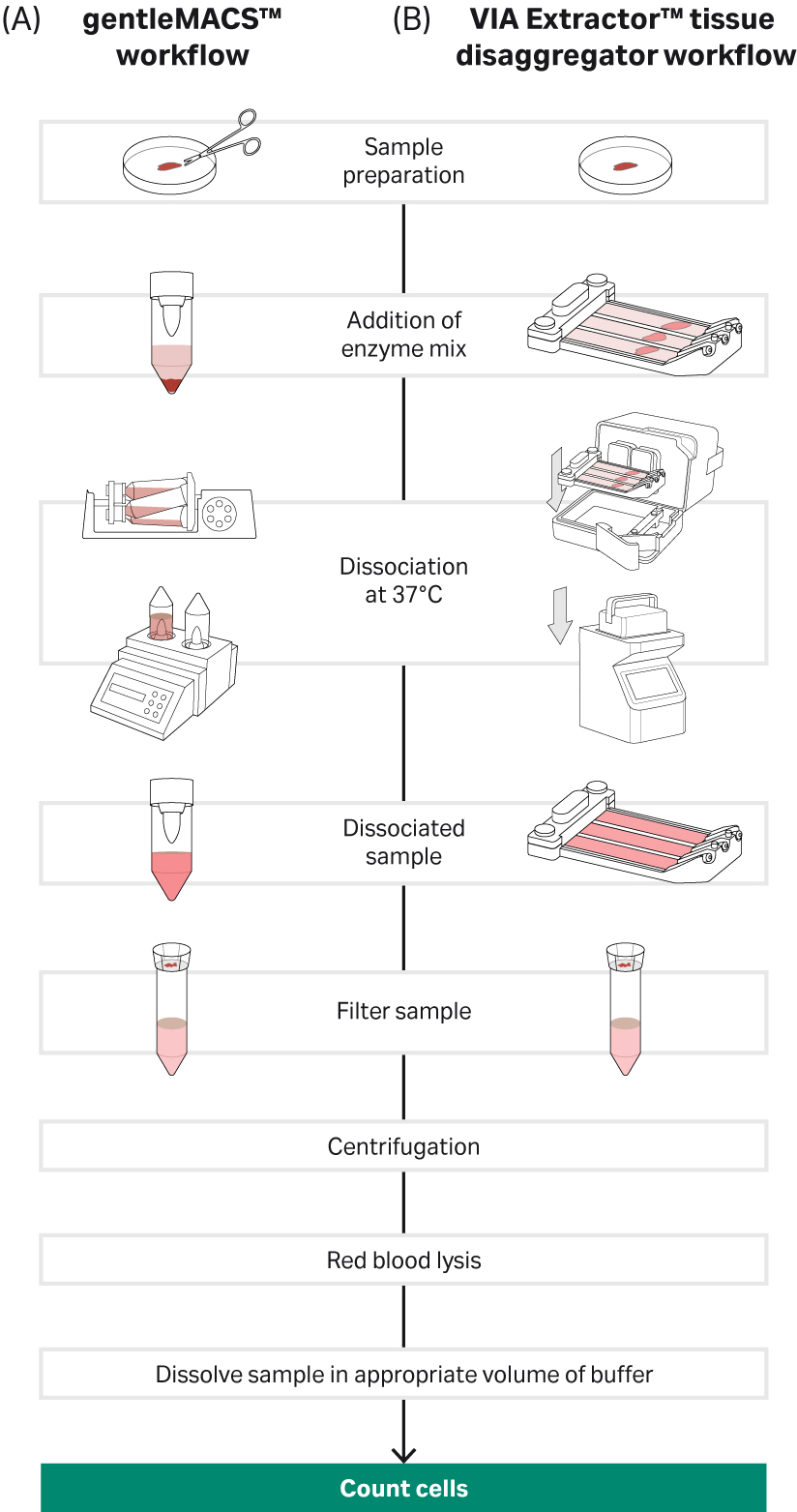
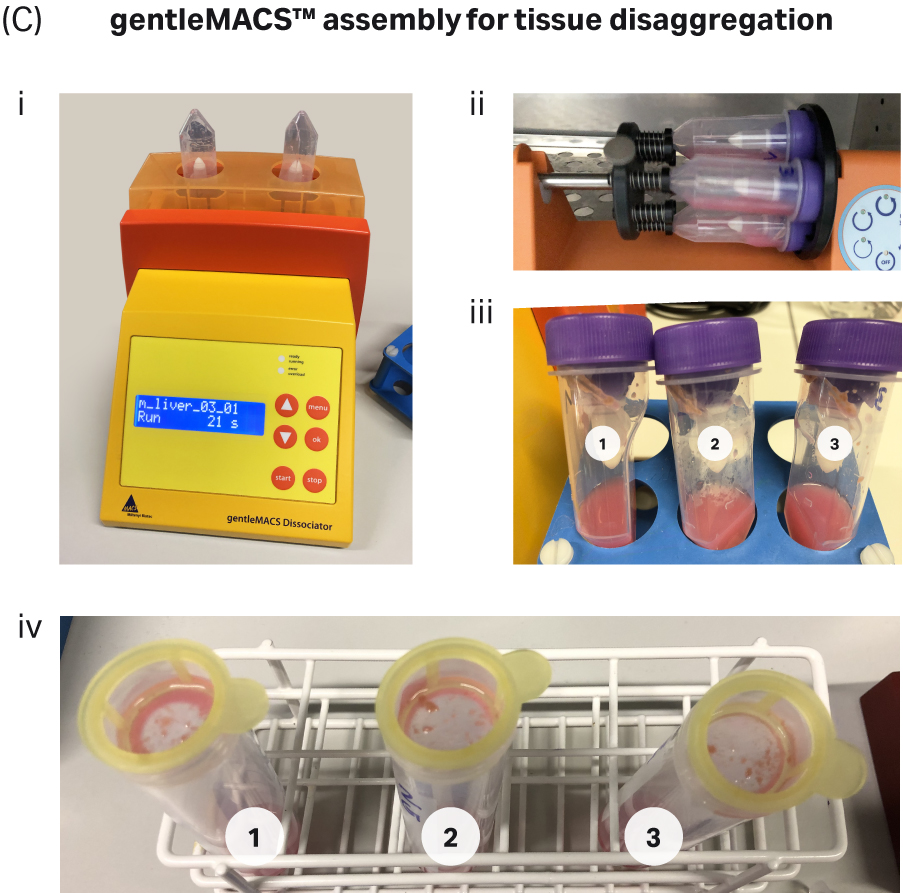
For the gentleMACS dissociator, liver tissue was washed with pre-warmed Dulbecco's Modified Eagle Medium (DMEM) before adding to each C tube containing the enzyme cocktail from the Liver Dissociation Kit (Miltenyi Biotec). The C tube was placed on the gentleMACS™ dissociator and the defined program for liver run for 37 seconds, following which the sample was incubated at 37°C for 30 mins on the MACSmix™ Tube Rotor (Miltenyi Biotec). Following incubation, each C tube was again placed on the gentleMACS™ dissociator and the second defined program for liver was run to generate the cell suspension. Following filtration of the samples, tissue debris was clearly visible on the cell strainers indicating that the tissue had not completely disaggregated (Fig. 2C iv).
For the VIA Extractor™ tissue disaggregator, liver tissue was washed with pre warmed DMEM before insertion into the Omics pouch. After sealing, the same enzyme cocktail as used for the gentleMACS™ protocol was added to the pouch via the syringe port. The Omics pouch was then placed into the VIA Extractor™
tissue disaggregator and digestion was carried out for 10 minutes at 37°C. The VIA Extractor™ tissue disaggregator completely dissociated the liver tissue in 10 minutes, with no debris evident on the cell strainers following filtration of the samples (Fig 2D ii and 2D iii).
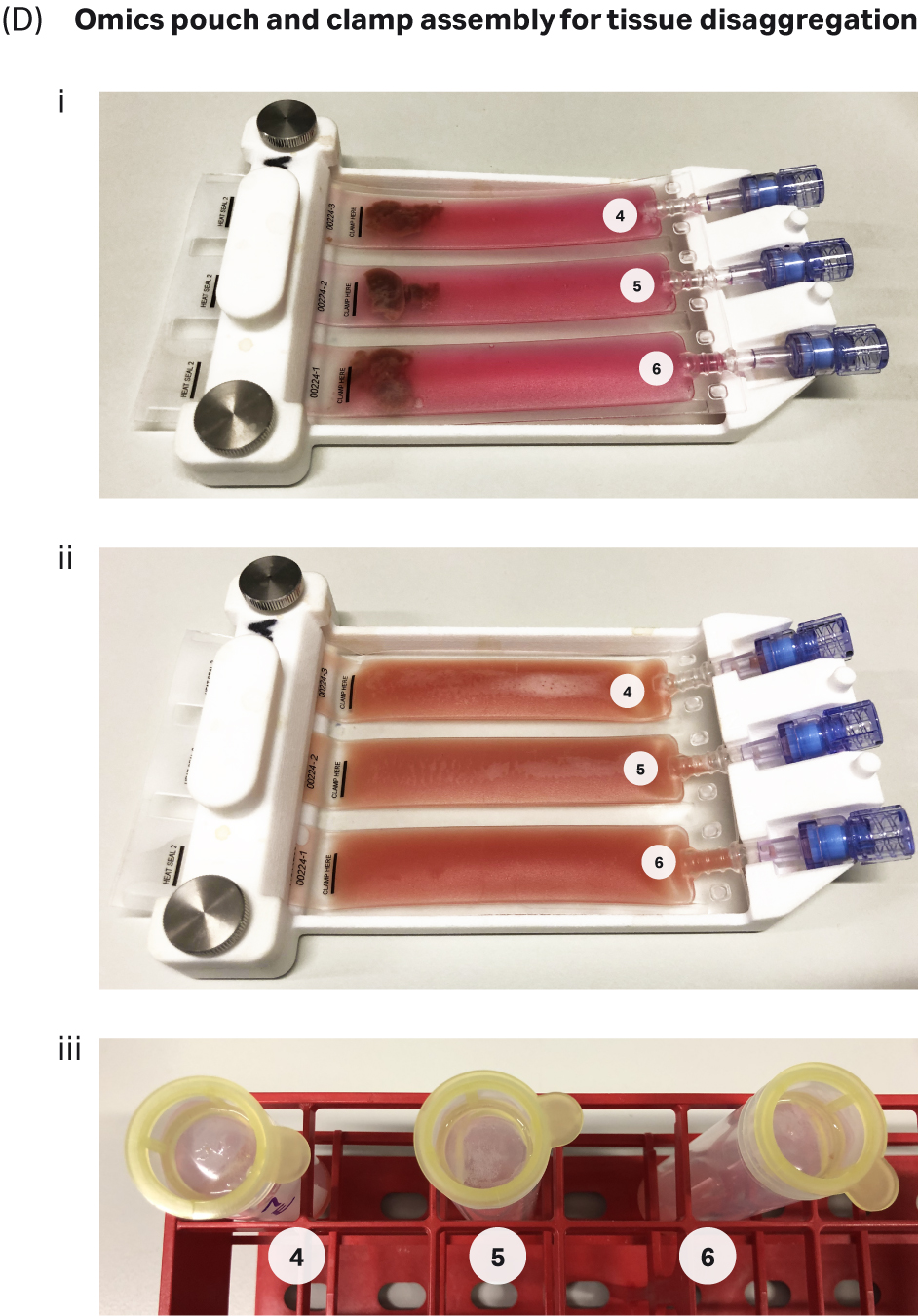
Fig 2. Tissue dissociation workflow for the gentleMACS™ and VIA Extractor™ tissue disaggregator: A) Complete workflow for gentleMACS™ dissociation method. B) Complete workflow for VIA Extractor™ tissue disaggregator dissociation method. C) gentleMACS™ C tubes were used (C-i) along with MACSmix™ Tube Rotor (C-ii) to disaggregate liver tissue. Single-cell suspension was obtained (C-iii) after the 30 minute incubation. Cells were passed over pre wet 100 µM cell strainers (C-iv). Cells were then passed over pre-wet 100µM cell strainers (D-iii) to filter out any undigested tissue material and debris. D) VIA Extractor™ tissue disaggregator assembly for tissue disaggregation. (D-i) Samples with enzyme mix in Omics assembly. (D-ii) Samples following 10 minutes disaggregation at 37°C. (D-iii) Cells were then passed over pre-wet 100µM cell strainers (D-iii) to filter out any undigested tissue material and debris.
Following dissociation, all samples (both methods) were centrifuged at 300 × g for 10 minutes at 4°C. Supernatants were carefully discarded, and red blood cells removed using the RBC Lysis Buffer (Miltenyi Biotec) according to the manufacturer’s protocol. Finally, each sample was carefully diluted to a working concentration of 800 cells/µL. The samples were then immediately processed by the Earlham Institute, Norwich, UK. Following quality evaluation, single cells produced from each system were processed using the Chromium™ controller (10x Genomics™) and libraries were sequenced using the NovaSeq™ 5000 platform (Illumina).
Results
The VIA Extractor™ tissue disaggregator from Cytiva produced higher yields of single cells in suspension with better viability, when compared to the gentleMACS™ dissociator.
Cell counts were consistently higher for samples generated using the VIA Extractor™ tissue disaggregator compared with gentleMACS™. Cell viability was also higher on samples prepared using VIA Extractor™ tissue disaggregator (samples 4-6, 72% ± 7% cell viability) versus gentleMACS™ cell dissociator (samples 1-3, 52% ± 3.6% cell viability) (Fig 2). This reduced efficiency is consistent with the observation that a considerable amount of undigested liver tissue debris remained on the cell strainer following use of the gentleMACS™ system (Fig 2).
Following dissociation, all samples were counted using automated TC20 cell counter and confirmed with the Haemocytometer. Results are shown in Table 4. Samples 1-3 were obtained using the gentleMACS™ dissociator while samples 4-6 were obtained using VIA Extractor tissue disaggregator. Viability and yield obtained using the VIA Extractor™ tissue disaggregator were significantly higher than for the gentleMACS™. Cell counts as determined after sequencing from Cell Ranger software were significantly higher when using the VIA Extractor™ tissue disaggregator. Note that all samples were normalized to 800 cells/µL prior to partitioning, suggesting that more cells survive the process when prepared using the VIA Extractor™ tissue disaggregator.
Table 4. Cell viability and yield
| Sample | Method | Total cells/mL | Live count cells/mL | % viability cells/mL | Haemocytometer cells/mL |
|---|---|---|---|---|---|
| 1 | gentleMACS™ | 4.64 x 106 | 2.59 × 106 | 56% | 2.2 × 106 |
| 2 | gentleMACS™ | 6.13 x 106 | 3.01 ×106 | 49% | 2.9 ×106 |
| 3 | gentleMACS™ | 5.35 x 106 | 2.75 × 106 | 51% | 2.3 × 106 |
| 4 | VIA Extractor™ tissue disaggregator | 9.94 x 106 | 5.30 × 106 | 77% | 4.9 × 106 |
| 5 | VIA Extractor™ tissue disaggregator | 1.07 x 107 | 5.81 × 106 | 75% | 5.1 × 106 |
| 6 | VIA Extractor™ tissue disaggregator | 1.71 x 107 | 6.71 × 106 | 64% | 5.9 × 106 |
Single-cell sequencing data generated via 10x Chromium platform reveals that single-cells suspension preparations obtained using VIA Extractor™ tissue disaggregator produce more cells and lower levels of cellular stress than the gentleMACS™.
To better understand the impact of the two tissue dissociation methods (VIA Extractor™ tissue disaggregator vs gentleMACS™) on the single-cell suspensions, single cell RNA sequencing was carried out on all samples described above. The aim was to investigate any transcriptional alterations induced on the cells by the different dissociation methods.
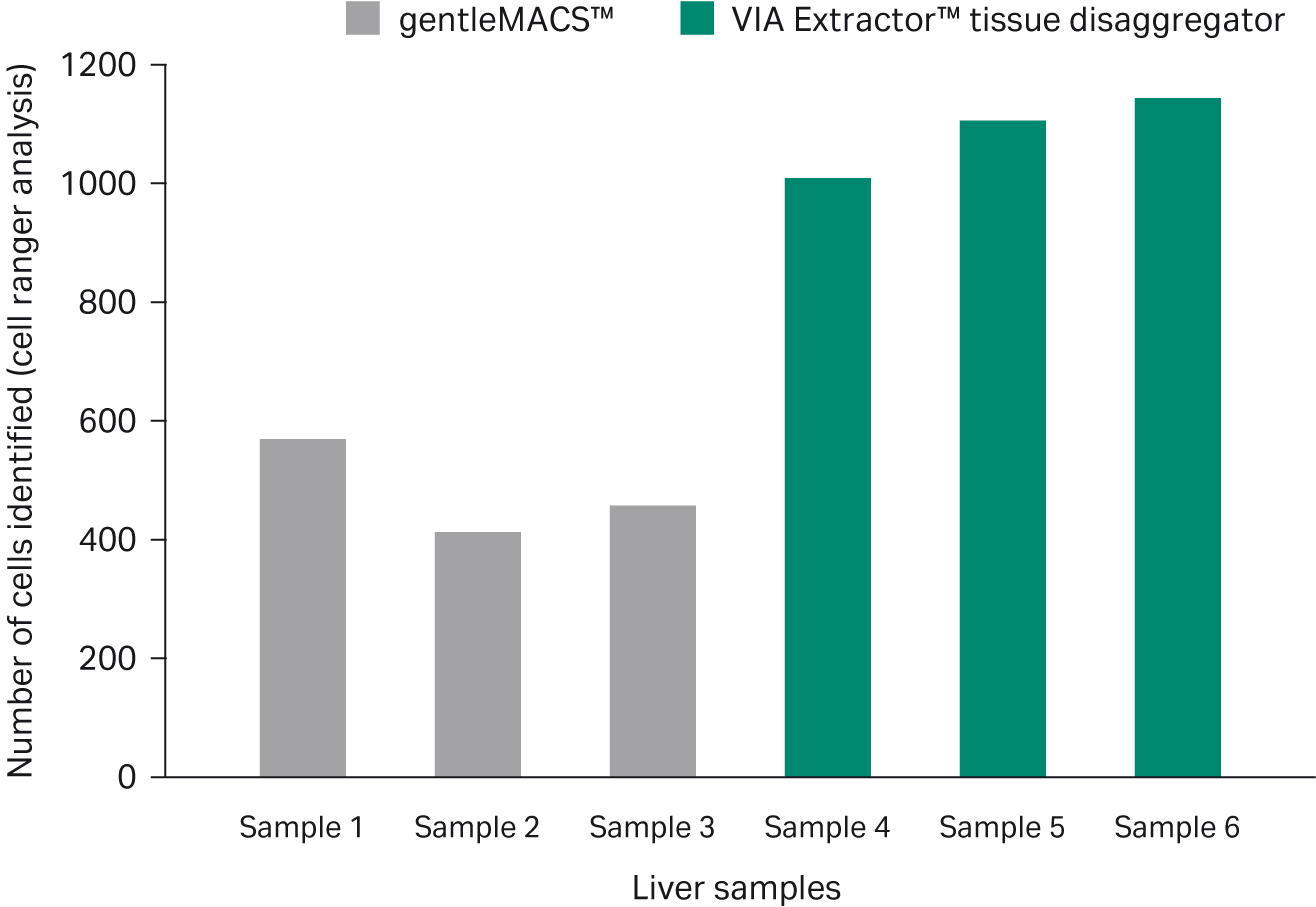
Although all samples were adjusted to 800 cells/µL prior to loading onto the Chromium controller, post sequencing quality control checks using Cell Ranger software revealed that the estimated number of recovered cells was higher in samples processed using the VIA Extractor™ tissue disaggregator (1086.3 ± 69.6 cells) compared to gentleMACS™ (479.3 ± 80.8 cells) (Fig 3). In addition, the fraction of reads in cells, was higher in samples generated using the VIA Extractor™ tissue disaggregator (59.7% ± 5.7%) compared to gentleMACS™ (49.8% ± 7.4%). Importantly, the fraction of reads in cells obtained using VIA Extractor™ tissue disaggregator is similar to the 58.4% reported for human liver data deposited on 10xQC website (https://10xqc.com/). Overall, these sequencing results indicate that the quality of the cells generated using the VIA Extractor™ tissue disaggregator is superior to gentleMACS™ for both viability of cells and number of cells recovered.
Fig 3. Cell viability. Cell counts as determined after sequencing from Cell Ranger software were significantly higher when using the VIA Extractor™ tissue disaggregator. Note that all samples were normalized to 800 cells/µL prior to partitioning, suggesting that more cells survive the process when prepared using the VIA Extractor™ tissue disaggregator
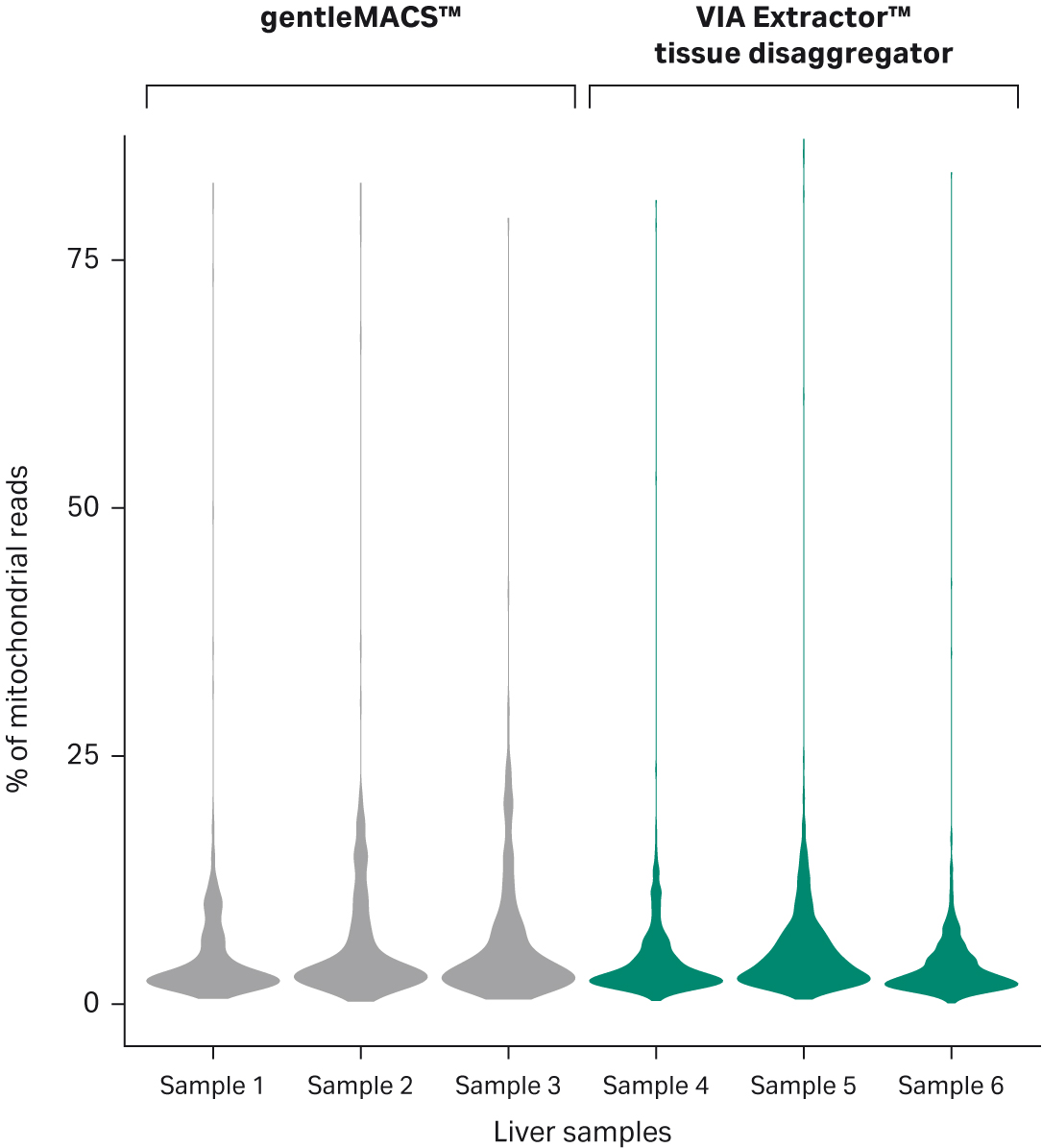
Further analysis of the data indicates that the samples were not overly enriched for mitochondrial transcripts, suggesting that overall, the sequenced cells were viable and not overtly stressed or dying (Fig 4).
Fig 4. Distribution of percentage of mitochondrial transcripts per sample. The percentage distribution of mitochondial content in samples confirms that the cells were viable when subjected to 10x Genomics workflow. Samples 1–3 were processed using gentleMACS™, while samples 4–6 were processed using the VIA Extractor™ tissue disaggregator.
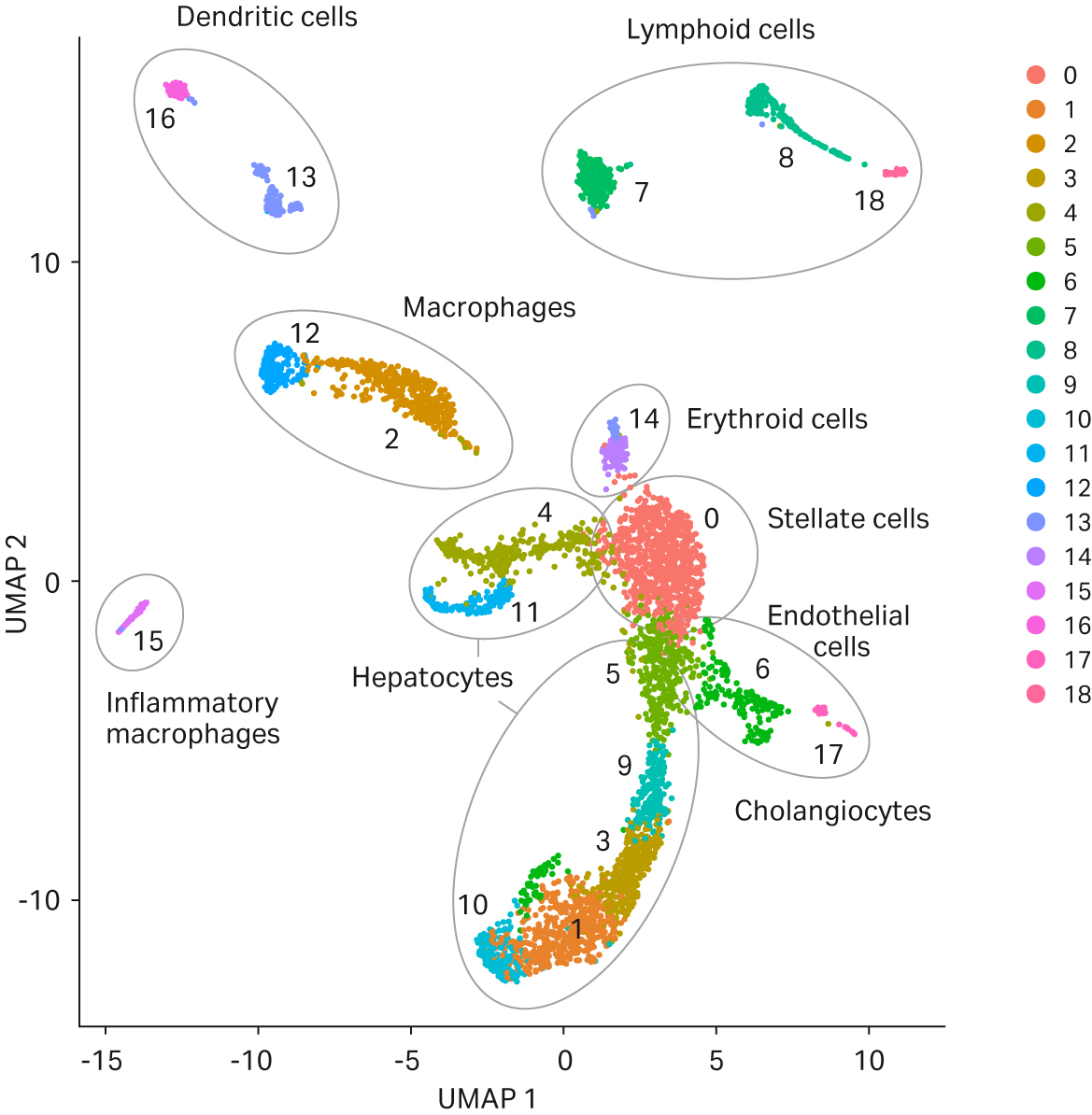
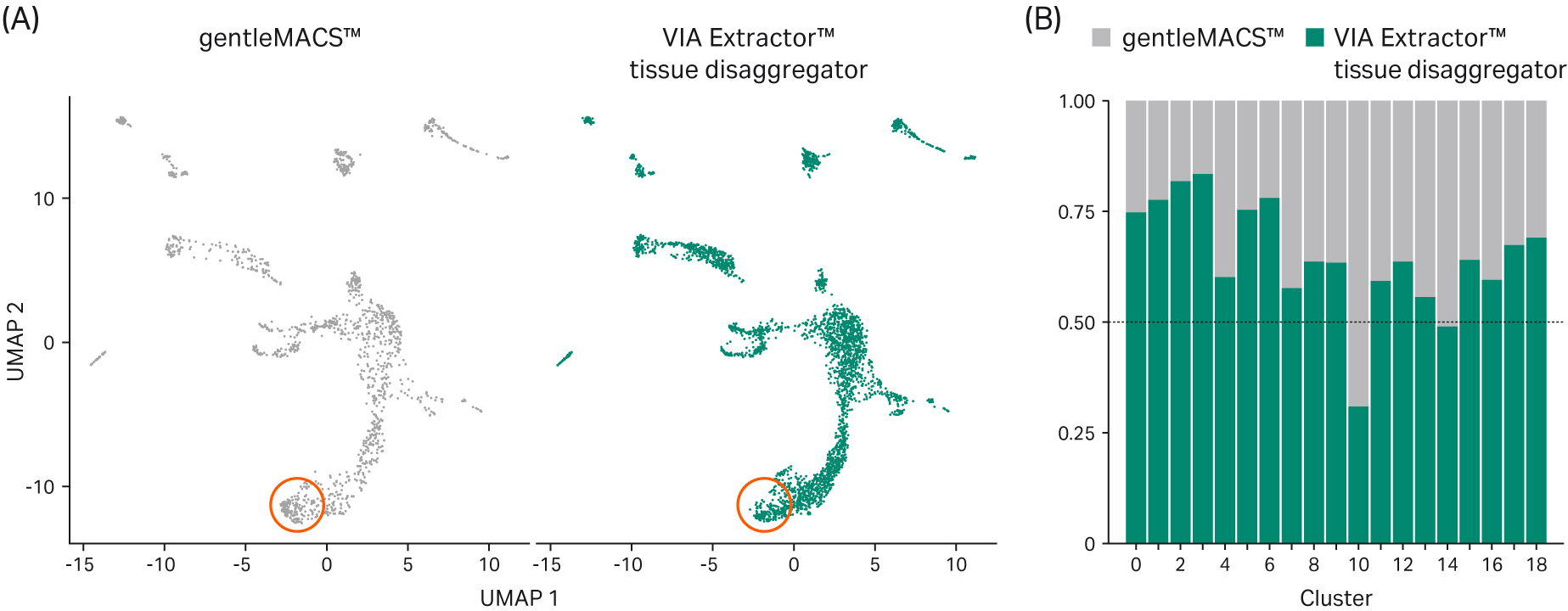
Although the same clusters and cell types were identified using both tissue dissociation methods, on average, twice as many cells from the VIA Extractor™ tissue disaggregator sample set were represented in the analysis compared with gentleMACS™ (Fig 6).
Fig 5. UMAP clustering of cell samples. Following sequencing, 19 clusters were automatically generated for both sample sets by Seurat analysis and cell types assigned
Interestingly, deeper analyses of the data revealed that the proportion of cells within each cluster was consistently higher in the sample generated using the VIA Extractor™ tissue disaggregator, with the exception of cluster 10, where the proportion of cells was higher in samples generated by gentleMACS™ (Fig 6B).
Fig 6. UMAP clustering of cells and differential abundance of cells. (A) UMAP clustering of all cells by sample set (gentleMACS™ and VIA Extractor™ tissue disaggregator). While the clustering is similar, there are significantly more cells in each cluster from tissue processed using the VIA Extractor™ tissue disaggregator. (B) Proportional representation of cells from both sample sets in each of the clusters identified in Figure 5 indicate that for samples processed using the VIA Extractor™ tissue disaggregator there are significantly more cells present in most clusters with the exception of cluster 10 which is higher for samples processed using the gentleMACS™ system
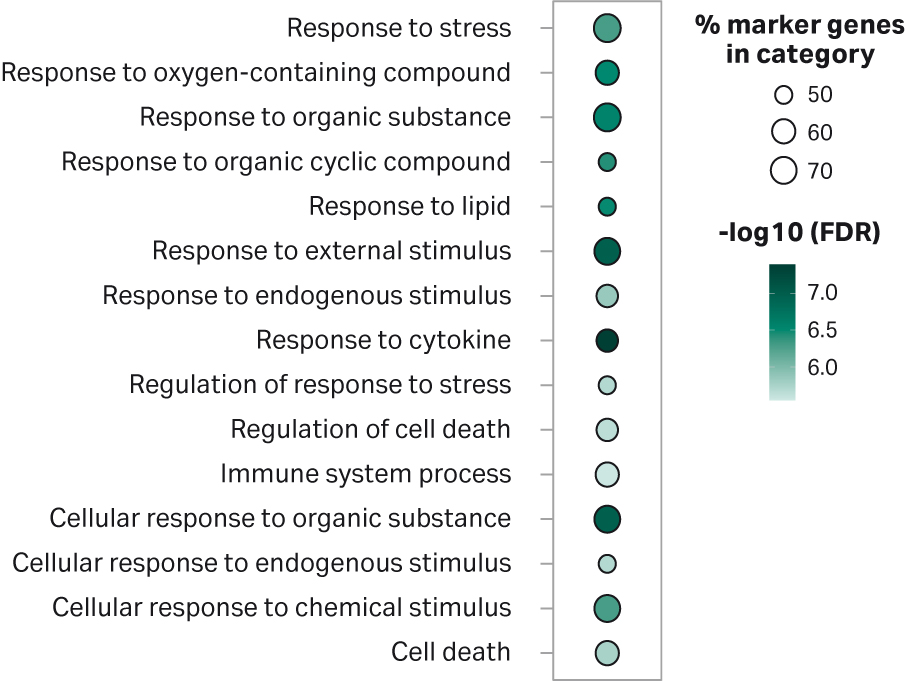
To identify the nature of the cells overrepresented in cluster 10 of the samples processed using gentleMACS™, a gene ontology (GO) analysis was performed on the top 20 most highly expressed genes that were detected in this cluster. This analysis revealed that the cells in cluster 10 were highly enriched in active biological processes that are associated with cellular stress and cell death (Fig 7). This observation is consistent with the fact that fewer cells passed the threshold for number of reads per cell as per 10x Genomics’ recommendation than for the VIA Extractor™ tissue disaggregator. This suggests that the single cells generated by gentleMACS™ are more fragile, being prone to break before or during the processing of the sample for sequencing.
Fig 7. Gene Ontology Analysis for the top 20 gene markers detected in cells within cluster 10. The list of genes was analyzed using ShynyGO v0.61 , which provides a list of functional categories and its respective enrichment FDRs (2). The results were plotted in R (v4.0.2) using a custom script. The top 15 more significant functional categories are shown. The percentage of marker genes in category represents the number of genes from the top 20 gene list found to belong to any individual category.
Conclusion
Single-cell RNA sequencing methodologies are providing deep insights into the roles of individual cells in the context of tissues, organs and even whole organisms, helping to unravel the molecular networks underlying both embryonic development and disease. The first step in many such workflows is the dissociation of tissue samples into single-cell suspensions prior to sequencing. It is well understood that the health and viability of cells following dissociation can be key to achieving the best possible results, so choosing the lowest impact technology for generation of single-cell suspensions should be a key consideration when setting up any single-cell omics workflow. Data from this analysis showed that the VIA Extractor™ tissue disaggregator from Cytiva provided a higher yield of viable mouse liver cells in a significantly shorter time period when compared to the gentleMACS™ dissociator. This is important for reducing process bias. UMAP clustering was similar between systems but showed that twice as many cells were represented in the sequencing data across nearly all clusters in single-cell suspensions generated using the VIA Extractor™ tissue disaggregator. Further gene ontology analysis indicated the presence of significantly elevated numbers of cells in the gentleMACS™ samples that displayed transcription profiles associated with cell fragility.
- Sonya A. MacParland et al., (2018) Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nature Communications
- Ge et al. (2020) Bioinformatics, Volume 36, Issue 8, Pages 2628–2629
*For research use only. Not for diagnostic use.
*This data is based on a minimum of three independent experiments and/or replicate trials with the equal number of replicates in each experiment. All samples tested were treated equally (with the number of replicates being the same for all products tested in the comparison) and according to manufacturers’ protocol and recommendations. Data was collected at Biomedical Research Centre (BMRC), University of East Anglia, NR4 7TJ and Earlham Institute, Norwich Research Park, Colney Lane, Norwich, Norfolk, England, NR4 7UZ during 29th of July 2020 and is held at Cytiva, Sovereign House, Chivers Way, Histon, Cambridge CB24 9BZ (R&D Laboratory).