By Monika Seidel PhD, Senior Development Scientist, Genomics and Cellular Research, Cytiva
Introduction
Liquid biopsy-based diagnostics using blood-based analysis of circulating cell-free DNA (cfDNA) holds great promise for early cancer screening, detection of minimal residual disease (MRD), and evaluating response to treatment.
For blood-based cfDNA analysis, both plasma and serum have been shown to be compatible with molecular profiling applications. However, plasma has become the sample of choice for cfDNA-based biomarker detection. This trend is well reflected in the number of scientific reports, the fast-growing market of specialized anticlotting blood collection tubes, and current workflows implemented by diagnostic laboratories.
The preferential use of plasma can be predominantly attributed to the requirement for allowing the blood to clot in serum processing. Blood clotting is associated with considerable release of genomic DNA (gDNA) from blood cells, which can result in significant loss in mutation detection sensitivity. Additionally, the clotting requirement is difficult to standardize.
The Sera-Xtracta Cell-Free DNA Kit* (Cytiva) has been designed to effectively capture cfDNA with minimal co-purification of higher molecular weight DNA. This kit has already been demonstrated to give a distinct advantage in cfDNA analysis when using plasma samples collected in both standard blood collection tubes and specialized cfDNA blood collection tubes. This application note present results from research we made to verify the suitability of the Sera-Xtracta cfDNA Kit for efficient extraction of cfDNA with concomitant reduction in genomic DNA from serum samples.
Biorepositories worldwide contain huge numbers of serum samples which are indispensable for retrospective large cohort studies for new cancer biomarker discovery. The identification of new mutations driving cancer development and progression constitutes the foundation of personalized medicine. This field relies on the discovery of novel targets for the development of targeted therapies that inhibit the mutated version of a given protein involved in cancer An efficient extraction of cfDNA from serum with concomitant reduction in gDNA is a critical tool for facilitating use of these serum samples in cancer-related research.
1. Extraction of cfDNA from serum of healthy donors
We collected 1 mL of serum from two healthy donors, used the samples for extraction with the Sera-Xtracta cfDNA Kit, and compared with the MagMAX™ Cell-Free DNA Isolation Kit (ThermoFisher Scientific). Fragment distribution of extracted sample(s) was analyzed using capillary electrophoresis on the 2100 Bioanalyzer™ (Agilent), with the High-Sensitivity DNA Kit (Agilent) (Fig 1A). For cfDNA recovery estimation, samples were run on the 7500 DNA kit (Agilent) and the DNA concentration within the main cfDNA peak was calculated with 2100 software smear analysis tool for fragments between 130 bp -260 bp (Fig 1B).
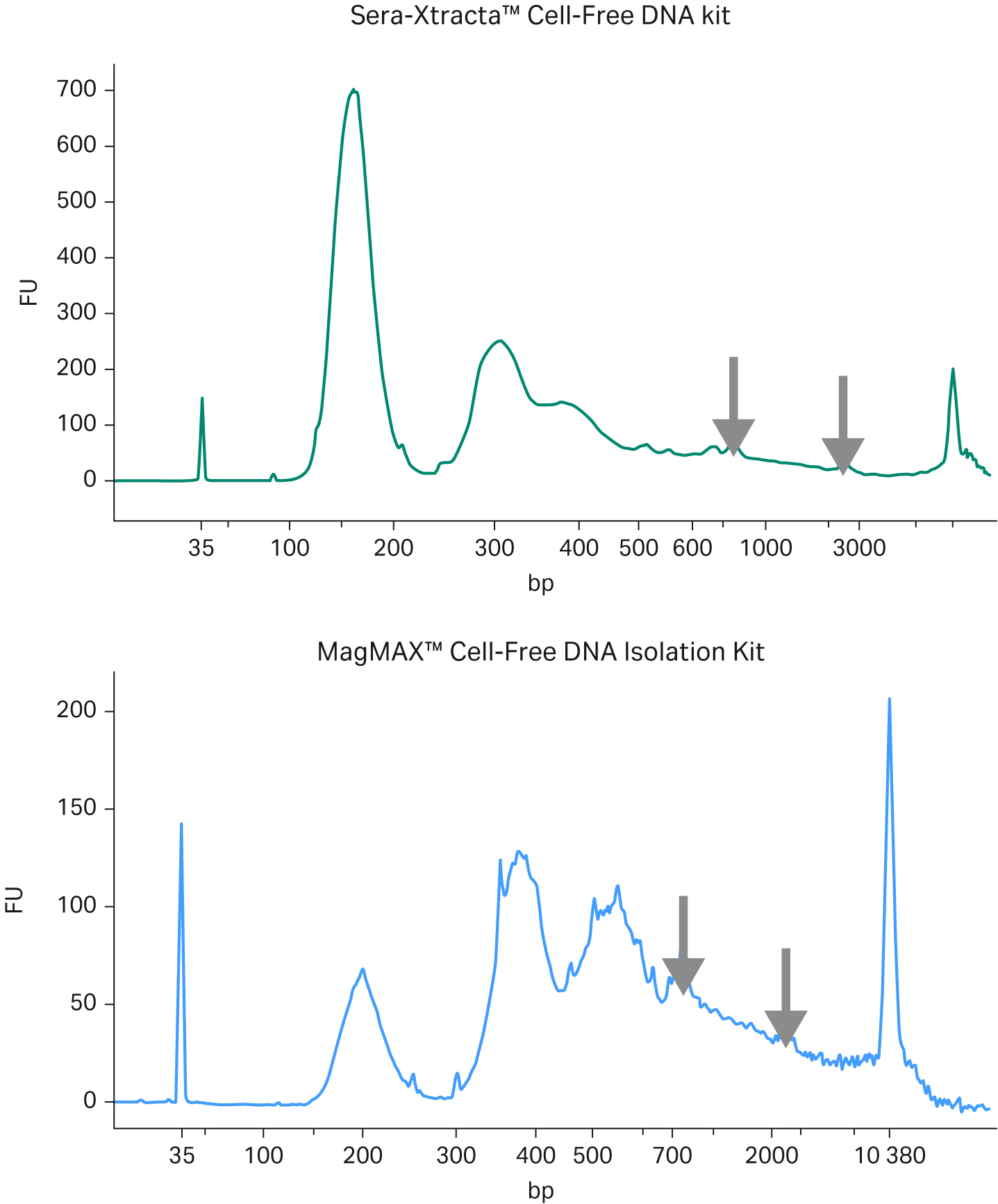
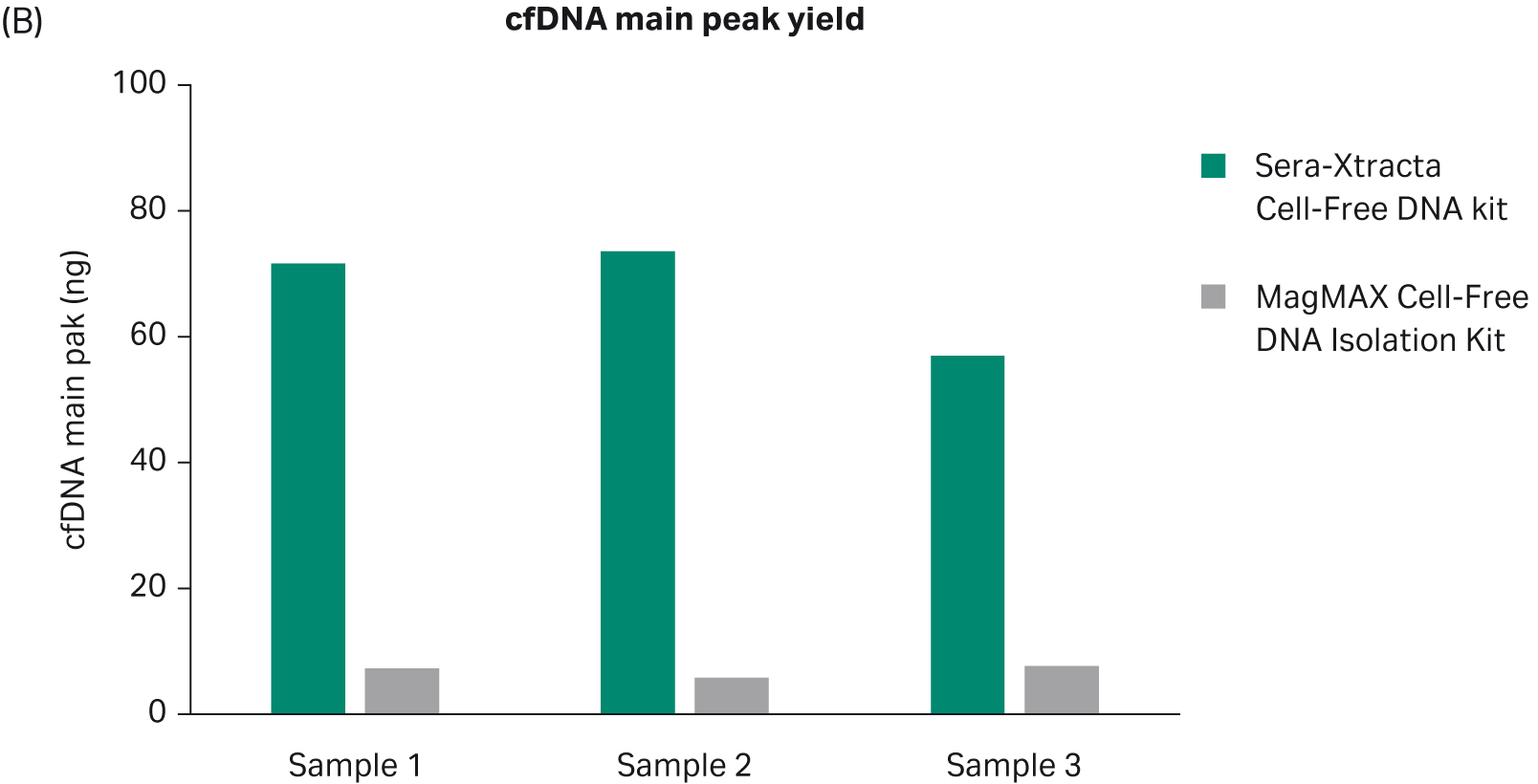
Fig 1. Extraction of cfDNA from serum samples from healthy donors; A) Representative electropherograms showing DNA fragment distribution obtained from healthy donor serum following cfDNA extraction using Sera-Xtracta Cell-Free DNA kit and MagMAX Cell-Free DNA Isolation kit. Extracted samples were analyzed on a 2100 Bioanalyzer using High-Sensitivity DNA Kit. Cell-free DNA is represented by a main peak centered around 170 bp, presence of high molecular weight (HMW) DNA represented by arrows, (B) Bar chart summarizing cfDNA recovery from serum calculated using smear analysis tool (2100 Expert software) for fragments between 130-260 bp following capillary electrophoresis on a 7500 DNA chip.
In agreement with the reported high level of gDNA in serum samples, representative electropherograms in Figure 1(A) show a significant amount of high molecular weight (HMW) DNA present in the eluted fraction for MagMAX Cell-Free DNA Isolation Kit, while the amount of gDNA carry-over is substantially reduced in the sample processed with Sera-Xtracta Cell-Free DNA Kit. More importantly, the yield of cfDNA is consistently and significantly higher (p < 0.01 based on paired t-test) in samples processed with the Cytiva product, unequivocally indicating the superior performance of Sera-Xtracta Cell-Free DNA Kit in serum samples.
2. Performance in targeted NGS: cancer biomarker detection in advanced stage cancer
Both non-small cell lung cancer (NSCLC) and colorectal cancer (CLC) are leading causes of mortality worldwide, with five year survival rate lower than 15% for the advanced stage (1,2). NSCLC treatment has undergone tremendous progress reflected in current clinical practice which involves routine testing for the presence of the genetic aberration in five genes most frequently mutated in NSCLC against which a targeted therapy has been approved (3). Unfortunately, targeted treatments for advanced CLC have not evolved as much and are currently restricted to vascular endothelial growth factor (VEGF) or epidermal growth factor (EGF) pathway inhibition, with decisions driven by the presence of additional predictive biomarkers (4).
Despite this significant shift from standard chemotherapy to precision medicine, which offers a prolonged progression-free survival and remarkable reduction in side-effects, there is still a significant number of patients who fail to respond to the first line targeted therapy. Patients who do initially respond to targeted therapy eventually develop secondary resistance that allows the cancer to bypass drug-mediated inhibition, leading to rapid disease progression. The discovery of novel cancer biomarkers and secondary resistance mutations is critical to enable further progress in the field that will ultimately translate into successful outcomes in more patients.
To further demonstrate the suitability of Sera-Xtracta Cell-Free DNA Kit for serum samples, we have evaluated its performance in next-generation sequencing (NGS) -based biomarker discovery workflow. For this purpose, we isolated cfDNA from 1 mL of serum from stage IV NSCLC and CLC patient samples with the Sera-Xtracta Cell-Free DNA Kit and the MagMAX Cell-Free DNA Isolation Kit and used in targeted NGS.
2.1 Cell-free DNA yield
We evaluated cell-free DNA yield as described above. Consistent with previous results, Sera-Xtracta Cell-Free DNA Kit outperformed MagMAX Cell-Free DNA Isolation Kit when used for extraction of cfDNA from serum (Fig 2).
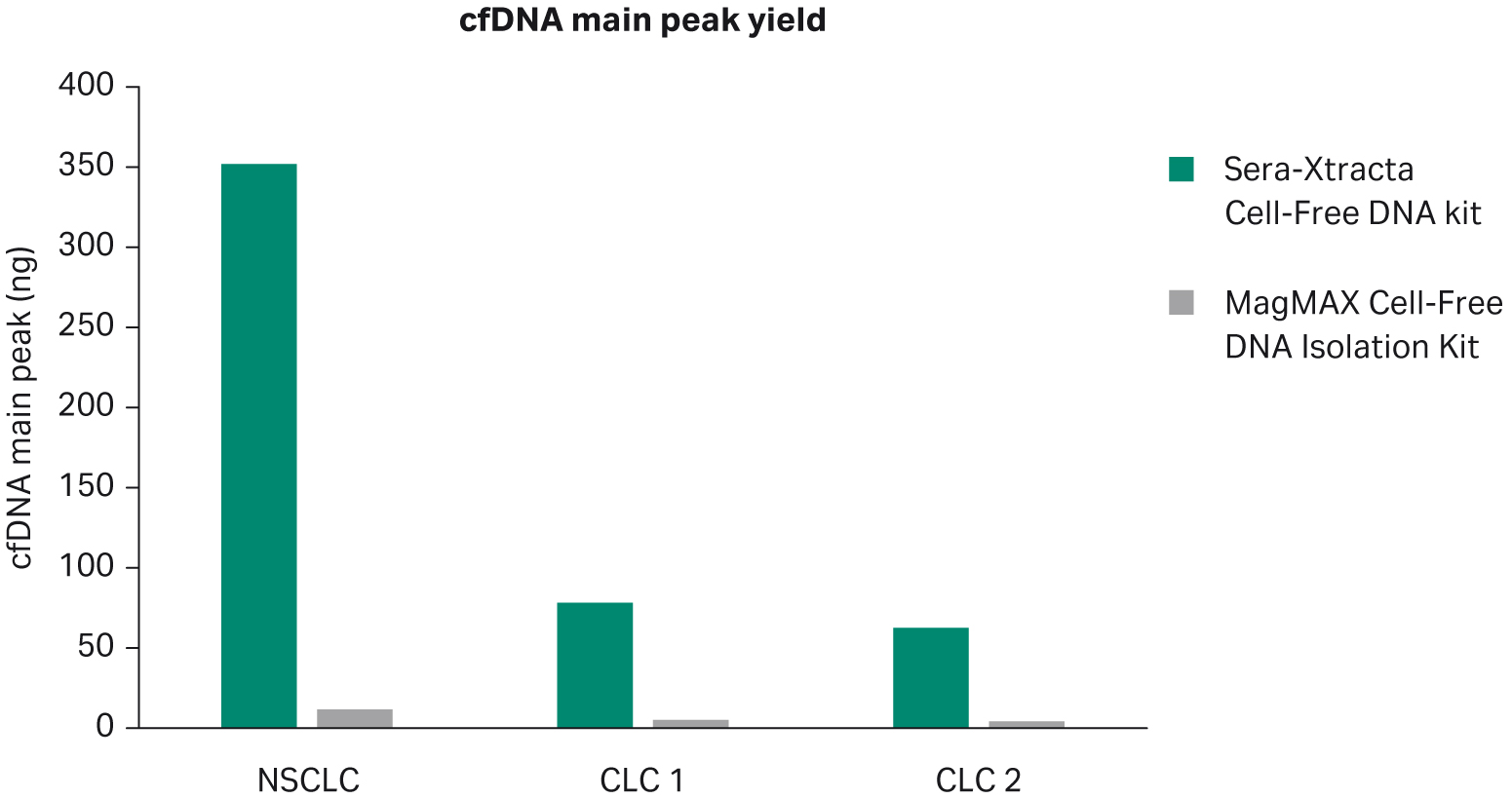
Fig 2. Extraction of cfDNA from serum samples from cancer patients. Bar chart summarizing cfDNA recovery following extraction with Sera-Xtracta Cell-Free DNA Kit and MagMAX Cell-Free DNA Isolation Kit. cfDNA yield calculated using a smear analysis tool (2100 Expert software) for fragments between 120-275 bp following capillary electrophoresis on a 2100 Bioanalyzer using the 7500 DNA Kit.
2.2 Targeted NGS library preparation
To prepare targeted NGS libraries using the hybridization-based approach, we used Trusight™ Tumor 170 RUO Kit (Illumina), which allows for the detection of somatic variants in 170 genes associated with cancer with > 95% sensitivity down to 5% mutation allele frequency (MAF). We used Qubit™ dsDNA HS Assay Kit (Invitrogen) to estimate total DNA concentration and adjusted DNA library input was according to manufacturer’s recommendations (input range 30-120 ng of total DNA). For all samples processed with the MagMAX Cell-Free DNA Isolation Kit, we prepared libraries using total remaining volume of extracted samples (12 µL: with input ranging from 20–95 ng). For the Sera-Xtracta Cell-Free DNA Kit, an equivalent of 120 ng was used. All nine libraries were pooled following bead-based normalization and run on the NextSeq™ (Illumina) instrument using High-Output Kit (Illumina) with paired-end reads of 101 bp.
2.3 Data analysis
For data analysis we used Trusight Tumor 170 BaseSpace App version 2.0.0 (Illumina) using BWA-MEM alignment to the human hg19 genome. App output files were filtered for all single nucleotide variant (SNV) in the protein coding sequence. In the absence of matched normal tissues control (e.g. blood gDNA), germline mutations were filtered out assuming that any call with the frequency range between 45%–55% is highly likely to represent a germline heterozygote and any call with MAF > 95% corresponds to a germline homozygote. The remaining SNVs which fulfilled sensitivity criteria (i.e. > = 5% MAF with minimum of 250x coverage) were considered potential somatic mutations.
2.4 Results
All samples were successfully sequenced; however, substantial differences in the quality of NGS data were evident, as shown in Table 1. Extracted cfDNA from Sera-Xtracta Cell-Free DNA Kit yielded libraries in which at least 98% of targets reached the minimum required sequencing depth (250x) for all samples, while target coverage in samples processed with the MagMAX Cell-Free DNA Isolation Kit was highly variable, and in one instance as low as 28.5% (p < 0.01 for mean target coverage, as shown in Figure 3, A and B.). Similarly, library complexity was consistently higher for all samples extracted with the Cytiva Kit (p < 0.05 for unique reads as shown in Figure 3C). Observed compromised library quality can be attributed to lower extraction efficiency obtained with the MagMAX Cell-Free DNA Isolation Kit. The Cell-free DNA extraction method developed by Cytiva minimizes sample processing-induced DNA damage, a phenomenon that creates sequencing artifacts such as chimeric reads (5). Consequently, the presence of chimeric reads was only detected in samples processed with MagMAX cfDNA Isolation kit. The total absence of chimeric reads in samples processed with Sera-Xtracta cfDNA Kit provides users with a representative, unaltered source of nucleic acid material that adequately represents input sample.
Table 1. Sequencing metrics as reported by Trusight Tumor 170 BaseSpace App version 2.0.0 (Illumina)
| NSCLC | CLC 1 | CLC2 | ||||
|---|---|---|---|---|---|---|
| Sera-Xtracta Cell-Free DNA |
MagMAX cfDNA kit | Sera-Xtracta Cell-Free DNA |
MagMAX cfDNA kit | Sera-Xtracta Cell-Free DNA |
MagMAX cfDNA kit | |
| CHIMERIC READS (%) | 0 | 0.1 | 0 | 0.1 | 0 | 0.1 |
| EXON COVERAGE at 250X (%) | 98.4 | 87.6 | 98.8 | 31.1 | 98.9 | 98.8 |
| TARGET COVERAGE at 250X (%) | 98.2 | 86.1 | 98.7 | 28.5 | 98.8 | 98.5 |
| MEAN TARGET COVERAGE | 2519.3 | 498.8 | 2343.5 | 189.7 | 2730 | 1179.3 |
| UNIQUE READS (%) | 38.5 | 13.3 | 38.5 | 14.3 | 36.7 | 22.2 |
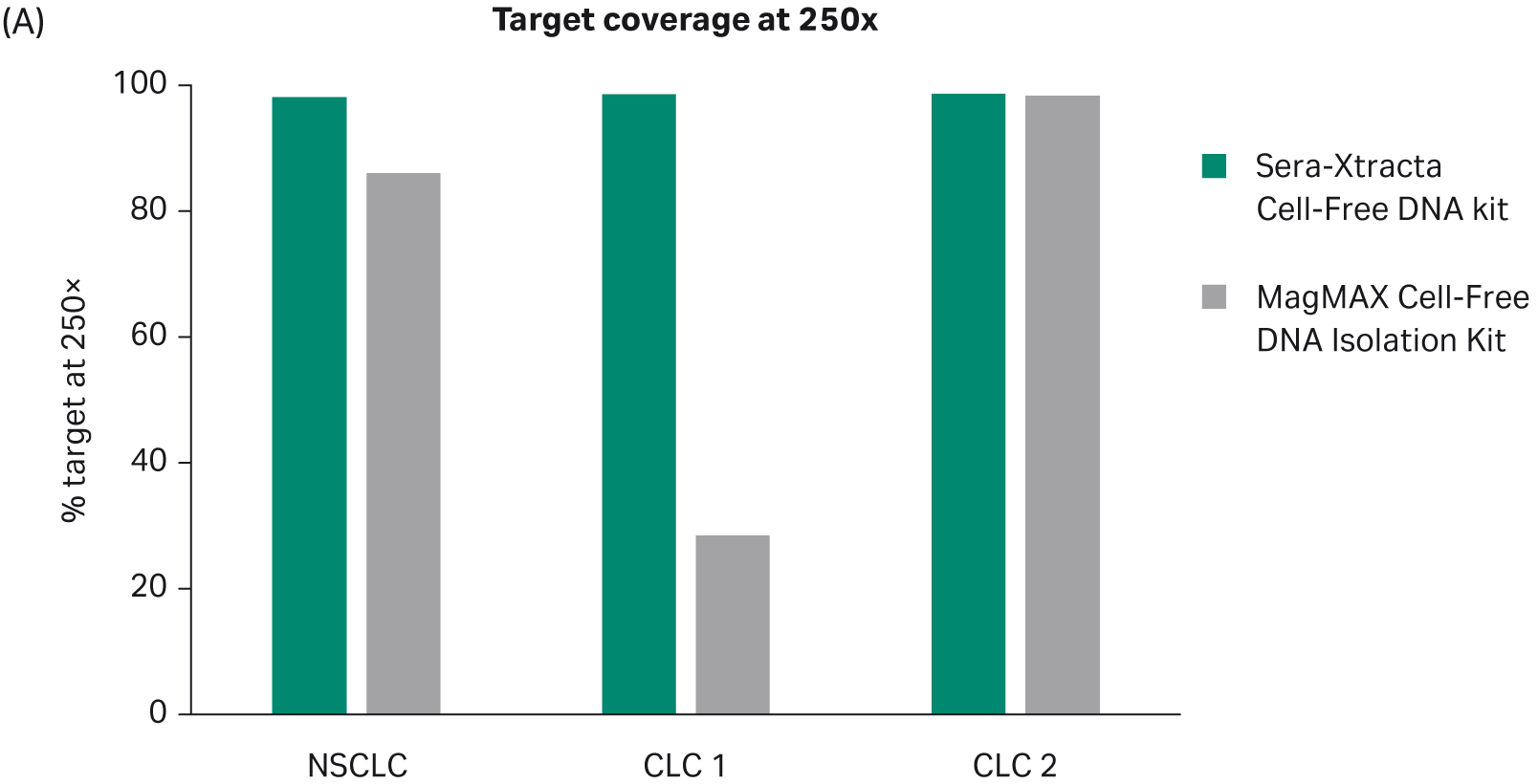
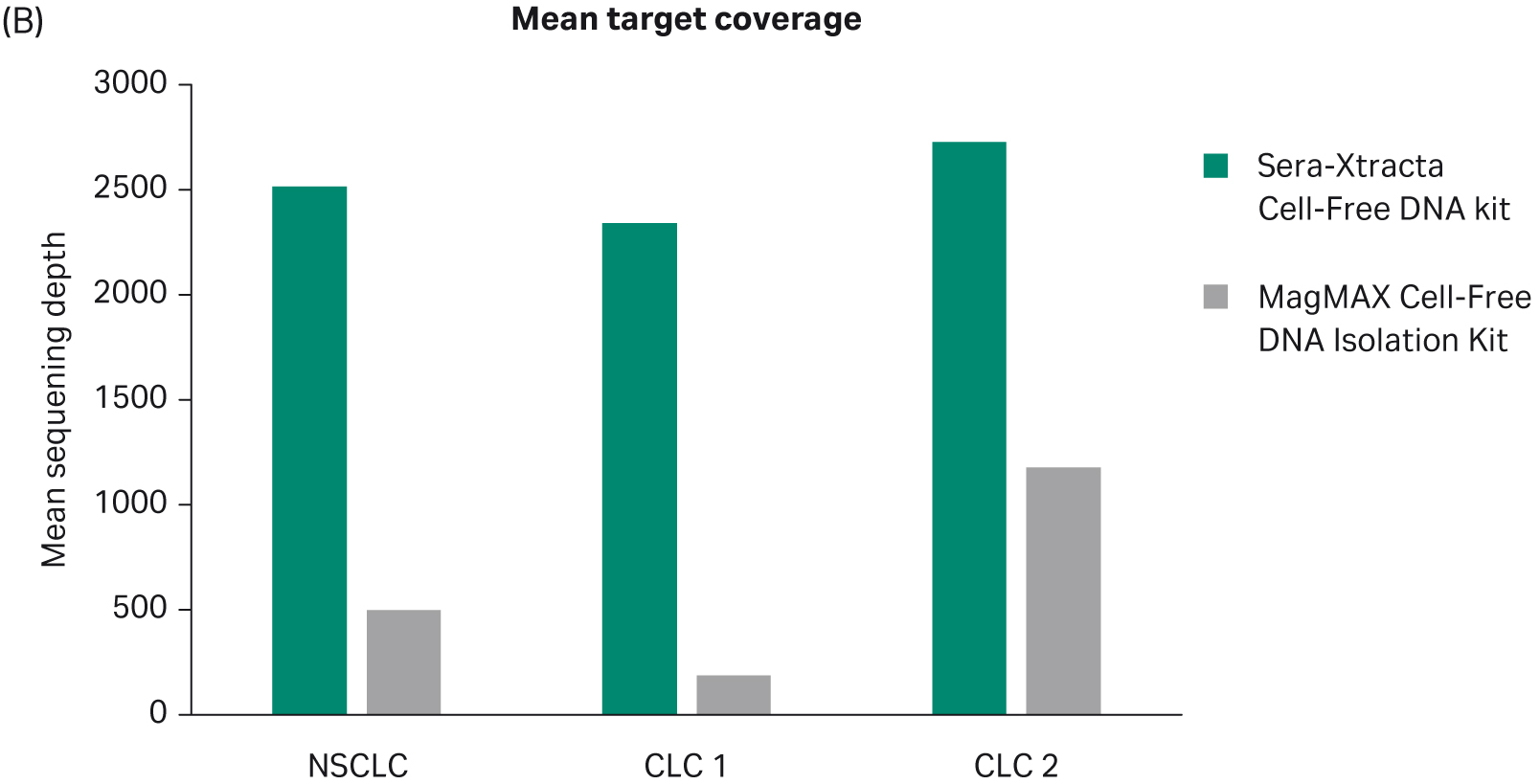
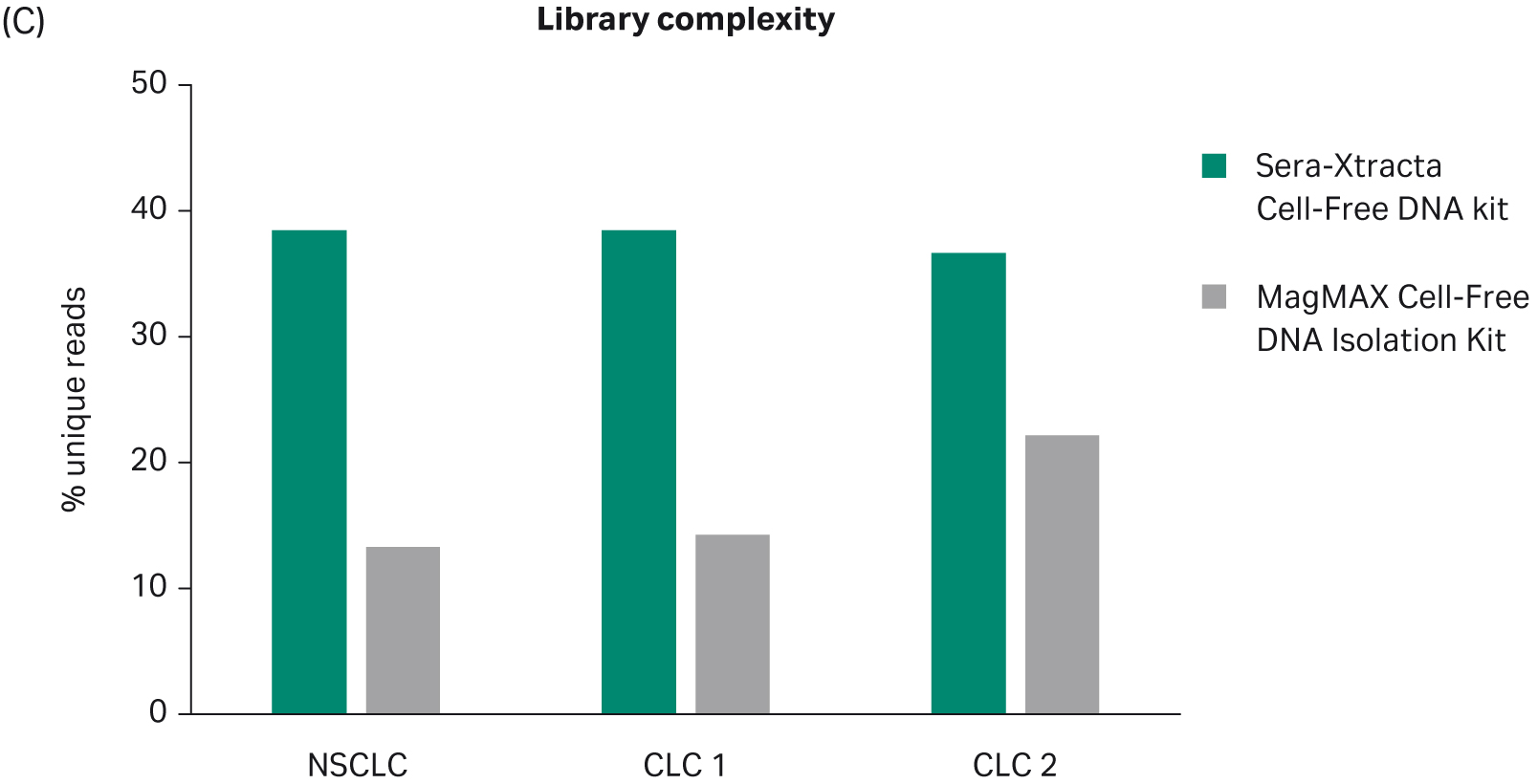
Fig 3. Summary of key NGS data metrics.
Concordant somatic SNV with potential clinical significance were detected in the serum of two patients (i.e. NSCLC and CLC 2) with comparable mutation allele frequency for both extraction methods (Table 2). However, it should be noted that, due to the much lower sequencing depth consistently obtained for samples processed with the MagMAX Cell-Free DNA Isolation Kit, the confidence of detection in those samples is compromised. In addition, due to the low cfDNA yield observed with the MagMAX Cell-Free DNA Isolation Kit, the entire eluted sample volume from the extraction step was used for the library preparation. This meant there was no more sample available for further confirmatory testing, if required.
Table 2. Somatic SNV detected in serum. Reported mutant allele frequency (MAF) represents the percentage of calls at a specific genomic position that are mutated over a cumulative number of calls at this position [(mutant/(mutant + wildtype)].
| Sera-Xtracta Cell-Free DNA Kit | MagMAX cfDNA kit | |||||
|---|---|---|---|---|---|---|
| Sample | Mutation | AA change | MAF | Sequencing depth | MAF | Sequencing depth |
| NSCLC | JAK2 | Val617Phe | 13% | 1725 | 15% | 921 |
| NRG1 | Met349Thr | 33% | 3479 | 35% | 1751 | |
| CLC 2 | IDH1 | Arg132His | 16% | 2264 | 13% | 342 |
| DNMT3A | Arg803Lys | 17% | 2328 | 12% | 664 | |
The V617F mutation in Janus kinase 2 (JAK2) gene identified in the serum of patient with advanced NSCLC is well documented in myeloproliferative disorders and targeted by two FDA approved drugs with proven clinical benefits for patients with advanced hematological malignancies (6). The role of JAK2 V617F mutation in NSCLC has been recognized only recently. It has been shown to predominantly co-exist with other mutations at a substantially lower MAF level suggesting a sub-clonal origin, and as such is a potential biomarker of tumor evolution (7). With growing evidence of the role JAK-STAT pathway in NSCLC, clinical trials of JAK inhibitors in patients with advanced NSCLCs are ongoing (https://clinicaltrials.gov).
The second mutation (NRG1: M349T) found in NSCLC serum samples has not been previously reported, and its pathogenicity and clinical utility remain unclear. Although rare, Neuregulin 1 (NRG1) mutations have been found in NSCLC; however, the predominant forms of genetic abnormalities involve gene fusions. The R132H mutation in isocitrate dehydrogenase (IDH1), identified in the patient with advanced CLC, is considered a direct driver of mutagenesis in glioma cancers and suggested to be an important biomarker of clinical and prognostic significance (8). Although, only a few cases of IDH-1/2 mutations have been reported in CLC, IDH mutations are starting to be recognized as potential drug targets in patients with advanced stages of the disease (9). This is reflected in ongoing clinical trials using IDH1 targeted inhibitors (NCT04584008, https://clinicaltrials.gov).
The second mutation identified in the serum of CLC patient (DNTM3A, R803K) is one of the most prevalent variants associated with acute myeloid leukemia (AML), and considered of negative prognostic value (10). The role of this mutation in CLC is not well understood, but aberrant DNMT3A-mediated DNA methylation in CLC has been widely reported, indicative of the role of this enzyme in gastric cancers (11).
3. Concluding remarks
Personalized medicine has revolutionized cancer treatment with unprecedented results in molecularly defined patient subgroups. This led to a dramatic shift in our approach to cancer treatment from cytotoxic chemotherapy targeting all rapidly dividing cells to patient specific biomarker driven therapeutic approaches with significantly improved survival and considerable reduction in systemic side-effects. The identification of novel mutations that might expose new therapeutic targets or prompt re-evaluation of already-approved drugs for new clinical indications is critical to drive progress in biomarker driven cancer therapy. In this respect, serum constitutes an important sample type that has great potential to facilitate the discovery of novel actionable biomarkers.
We have demonstrated that Sera-Xtracta Cell-Free DNA Kit efficiently extracts cfDNA from serum with superior extraction efficiency when compared to an alternative bead-based commercial product (MagMAX Cell-Free DNA Isolation Kit). It consistently yields cfDNA at sufficient quantity for sensitive downstream applications such as next-generation sequencing with as little as 1 mL of serum input. In targeted NGS, the Sera-Xtracta Cell-Free DNA Kit outperforms MagMAX Cell-Free DNA Isolation Kit in the quality of cfDNA extracted. This is reflected in the quality of the sequencing data such as higher library diversity, increased sequencing depth, and excellent target coverage, which are all necessary for confident SNV calls. In addition, in samples where a significant amount of gDNA is present, Sera-Xtracta cfDNA kit provides additional distinct advantage: it effectively reduces gDNA carry-over, ensuring maximum enrichment in cfDNA.
Learn more about Sera-Xtracta Cell-Free DNA Kit
* Sera-Xtracta cfDNA Kit is for research use only.
- Majem M, Remon J. Tumor heterogeneity: evolution through space and time in EGFR mutant non small cell lung cancer patients. Transl Lung Cancer Res. 2013;2(3):226-237. doi:10.3978/j.issn.2218-6751.2013.03.09
- Honma K, Takemasa I, Matoba R, et al. Screening of potential molecular targets for colorectal cancer therapy. Int J Gen Med. 2009;2:243-257. Published 2009 Dec 29. doi:10.2147/ijgm.s5158
- NCCN Foundation. Metastatic Lung Cancer - NCCN. National Comprehensive Cancer Network. https://www.nccn.org/patients/guidelines/content/PDF/lung-metastatic-patient.pdf. Published 2019.
- NCCN Foundation. NCCN Guidelines for Patients: Colon Cancer. National Comprehensive Cancer Network. https://www.nccn.org/patients/guidelines/content/PDF/colon-patient.pdf. Published 2018.
- Haile S, Corbett RD, Bilobram S, et al. Sources of erroneous sequences and artifact chimeric reads in next generation sequencing of genomic DNA from formalin-fixed paraffin-embedded samples. Nucleic Acids Res. 2019;47(2):e12. doi:10.1093/nar/gky1142
- Bose P, Verstovsek S. JAK Inhibition for the Treatment of Myelofibrosis: Limitations and Future Perspectives. Hemasphere. 2020;4(4):e424. Published 2020 Jul 21. doi:10.1097/HS9.0000000000000424
- Li SD, Ma M, Li H, et al. Cancer gene profiling in non-small cell lung cancers reveals activating mutations in JAK2 and JAK3 with therapeutic implications. Genome Med. 2017;9(1):89. Published 2017 Oct 30. doi:10.1186/s13073-017-0478-1
- Cohen AL, Holmen SL, Colman H. IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep. 2013;13(5):345. doi:10.1007/s11910-013-0345-4
- Fallah-Rad N, Bedard PL, Siu LL, et al. Association of isocitrate dehydorgenase-1 (IDH-1) mutations with elevated oncometabolite 2-hydroxyglutarate (2HG) in advanced colorectal cancer. J Clin Oncol. 2016;34(4_suppl):627-627. doi: 10.1200/jco.2016.34.4_suppl.627
- Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424-2433. doi:10.1056/NEJMoa1005143
- Skowronski K, Dubey S, Rodenhiser D, Coomber B. Ischemia dysregulates DNA methyltransferases and p16INK4a methylation in human colorectal cancer cells. Epigenetics. 2010;5(6):547-556. doi:10.4161/epi.5.6.12400