Here you will learn about how we optimized the purification of Adenovirus AdV5 using Ion Exchange Chromatography Columns and ÄKTA pure™ chromatography system.
Introduction
Adenovirus is evaluated as a vaccine delivery system in many preclinical and clinical studies for various infectious diseases (1). Recently, the advantage of simply altering the carried nucleic sequence to induce immune response has rapidly resulted in several Covid-19 vaccines using adenovirus as vector (2). Adenovirus is also explored as a potential viral vector for gene therapy and as an oncolytic virus. To enhance productivity and produce more effective and safer vaccines, several generations of recombinant adenovirus vectors have been developed (3). The most studied adenovirus vector is the first generation of recombinant adenovirus serotype 5 (AdV5).
This work shows the optimization of capturing AdV5 using anion exchange chromatography and evaluation of the core bead technology that combines size exclusion and binding properties for the polishing step. This results in a two-step purification of AdV5.
HiTrap™ Capto ™ Q ImpRes resin is a strong anion exchanger and was chosen for its small bead size, providing a larger surface area for virus binding and higher capacity.
Capto Core are chromatographic resins with an adsorbing core contained within an inert shell layer. The core of each bead is functionalized with ligands that are both hydrophobic and positively charged, resulting in a highly efficient multimodal binding of various impurities small enough to diffuse through the shell and enter the core. The shell of core beads do not have a singular pore size. It is rather a pore size distribution with an average cut-off corresponding to a 700 kDa globular entity for Capto™ Core 700 and 400 kDa globular entity for Capto™ Core 400 resin. They need to be evaluated to obtain the best impurity removal in the polishing step.
Optimization of capture of AdV5 on HiTrap™ Capto™ Q ImpRes resin
Dynamic binding capacity study of NaCl concentration in the sample and in binding buffer revealed that salt concentration as high as 300-450 mM was beneficial for capacity of AdV5 (not shown).
When the gradient was developed it was important to try to keep this as short as possible without co-elution of DNA. DNA is negatively charged and binds strongly to anion exchangers, and longer DNA sequences bind stronger than shorter DNA fragments. Earlier elution of the virus in the gradient reduces the risk of contamination of shorter DNA fragments in the virus fraction.
In figure 1, a gradient starting at 300 mM salt and ending at 700 mM in 11 column volumes (CV) is shown. The run also includes a CIP step with 1 M NaOH where tightly bound contaminants are cleaned from the resin. Virus was eluted in the second peak and total protein was reduced by 99% and hcDNA by 96% in the pooled fraction. The result was very promising with very good separation of virus particles and impurities. The method was further optimized to shorten the gradient and the results are shown in the next section.
Sample: adenovirus subjected to NFF (2 µm, 0.6 µm) and TFF (NMWC Mr 300000) in 20 mM Tris, pH 8.0 + 300 mM NaCl
Column: HiTrap™ Capto™ Q ImpRes 1 mL
Load: 2.9 mL
Flow rate: 0.1 mL/min
Binding buffer: 20 mM Tris, pH 8.0 + 300 mM NaCl
Wash: 3 CV of binding buffer
Elution: 20 mM Tris, pH 8.0 + 300 to 700 mM NaCl in 11 CV
System: ÄKTA™ pure 25
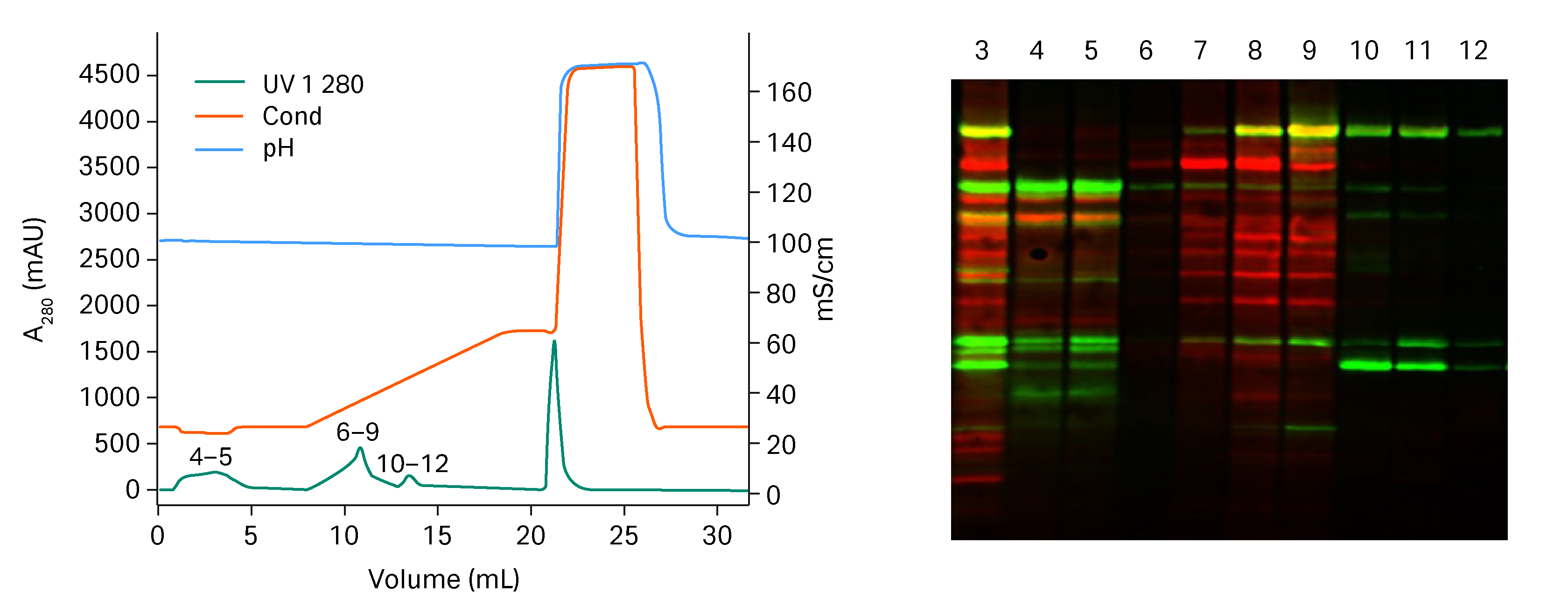
Fig 1. To the left, chromatogram from the Capto™ Q ImpRes run. To the right, Western blot results run where green equals viral proteins and red equals HCP. Numbers in chromatogram correspond to lanes in the Western blot. Lane 3 = starting material. Lane 4-5 = flow through and wash containing HCP and free viral proteins. Lane 6-9 = first elution peak containing mostly HCP. Lane 10-12 = second elution peak containing only viral proteins (confirmed with analytical anion exchange to be whole virus).
Evaluation of HiTrap™ Capto™ Core 400 and HiTrap™ Capto™ Core 700 column for virus polishing
Adenovirus is a medium size virus 90-100 nm and too large to enter the pores of either HiTrap™ Capto™ Core 400 or HiTrap™ Capto™ Core 700 column but the question was, which one will remove contaminants most efficiently. The comparison was performed with a sample that had been NFF/TFF treated and with higher impurity content than sample applied in a normal polishing step.
The flow through was collected and the fractions individually analyzed with western blot, see figure 2. The first fraction of HiTrap™ Capto™ Core 700 column, lane 3, does not contain any HCP. For HiTrap™ Capto™ Core 400 column it seemed that some large protein contaminants did not enter the pores and already the first fraction contained HCP, lane 6. HiTrap™ Capto™ Core 700 column was chosen as more suitable for the polishing step.
Sample: adenovirus subjected to NFF (2 µm, 0.6 µm) and TFF (NMWC Mr 300000) in 20 mM Tris, pH 8.0 + 300 mM NaCl
Columns: HiTrap™ Capto™ Core 700 1 mL, HiTrap™ Capto™ Core 400 1 mL column
Load: 10 mL
Flow rates: sample load 0.33 mL/min, CIP 0.5 mL/min
Buffer: 20 mM Tris, pH 8.0 + 300 mM NaCl
CIP: 1 M NaOH+ 27% 1-propanol, 5 CV with 30 min pause + 5 CV
System: ÄKTA pure™ 25
| Step | Volume | Buffer/material | Flow (mL/min) |
| Equilibration | 5 CV | 20 mM Tris, pH 8.0 + 300 mM NaCl | 1 |
| Sample load | 10 mL | TFF/NFF + 300 mM NaCl | 0.33 |
| Wash | 3 CV | 20 mM Tris, pH 8.0 + 300 mM NaCl | 1 |
| CIP | 5 CV | 1 M NaOH + 27% 1-propanol 30 min pause |
0.5 |
| CIP | 5 CV | 1 M NaOH + 27% 1-propanol | 0.5 |
| Re-equilibration | 10 CV | 20 mM Tris, pH 8.0 + 300 mM NaCl | 1 |
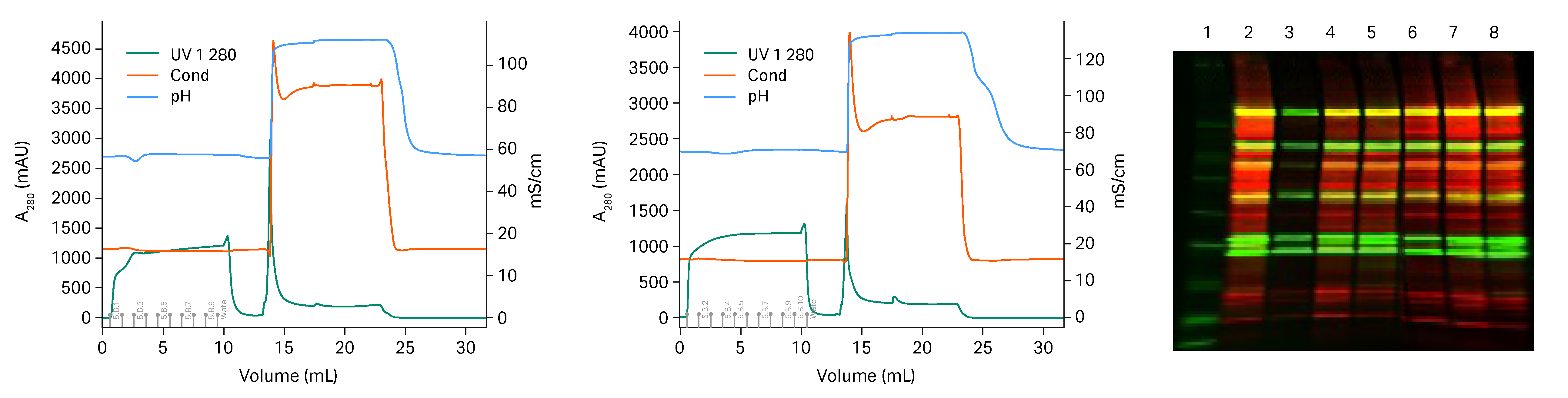
Fig 2. To the left, chromatogram from the HiTrap™ Capto™ Core 700 column run. In the middle, chromatogram from the HiTrap™ Capto™ Core 400 column run. To the right, Western blot results run where green equals viral proteins and red equals HCP. Numbers in chromatogram correspond to lanes in the Western blot. Lane 1=marker, Lane 2 = starting material. Lane 3-5 = flow through HiTrap™ Capto™ Core 700 column. Lane 6-8= flow through HiTrap™ Capto™ Core 400 column.
Two step purification of AdV5
Adenovirus subjected to NFF/TFF (24.8 mL) was loaded on HiScreen™ Capto™ Q ImpRes column with 450 mM NaCl in the binding buffer. In the optimized gradient, a wash step with 480 mM NaCl was added and the elution was performed in a very short gradient of 2.5 CV from 480 to 570 mM NaCl. HiTrap™ Capto™ Core 700 column was found to be best column for polishing and was loaded with 40 mL eluate from two pooled HiScreen™ Capto™ Q ImpRes column purifications, see Figure 4. The flow though was collected in fractions and the fractions were analyzed for total protein, genomic DNA and virus particles. A breakthrough of genomic DNA was found in the HiTrap™ Capto™ Core 700 column fractions after 26 CV load, where the fractions up to 26 CV load were pooled. Total protein and genomic DNA were under detection limit in the pool and the virus content was 1.3*1011 virus particles/mL and in total 3.4*1012 vp were purified.
Sample: adenovirus subjected to NFF (2 µm, 0.6 µm) and TFF (NMWC Mr 300000) in 20 mM Tris, pH 8.0 + 450 mM NaCl
Column: HiScreen™ Capto™ Q ImpRes
Load: 24.8 mL
Flow rate: 0.47 mL/min
Binding buffer: 20 mM Tris, pH 8.0 + 450 mM NaCl
Wash: 3 CV 20 mM Tris, pH 8.0 + 480 mM NaCl
Elution: 20 mM Tris, pH 8.0 + 480 to 570 mM NaCl in 2.5 CV
System: ÄKTA™ pure 150
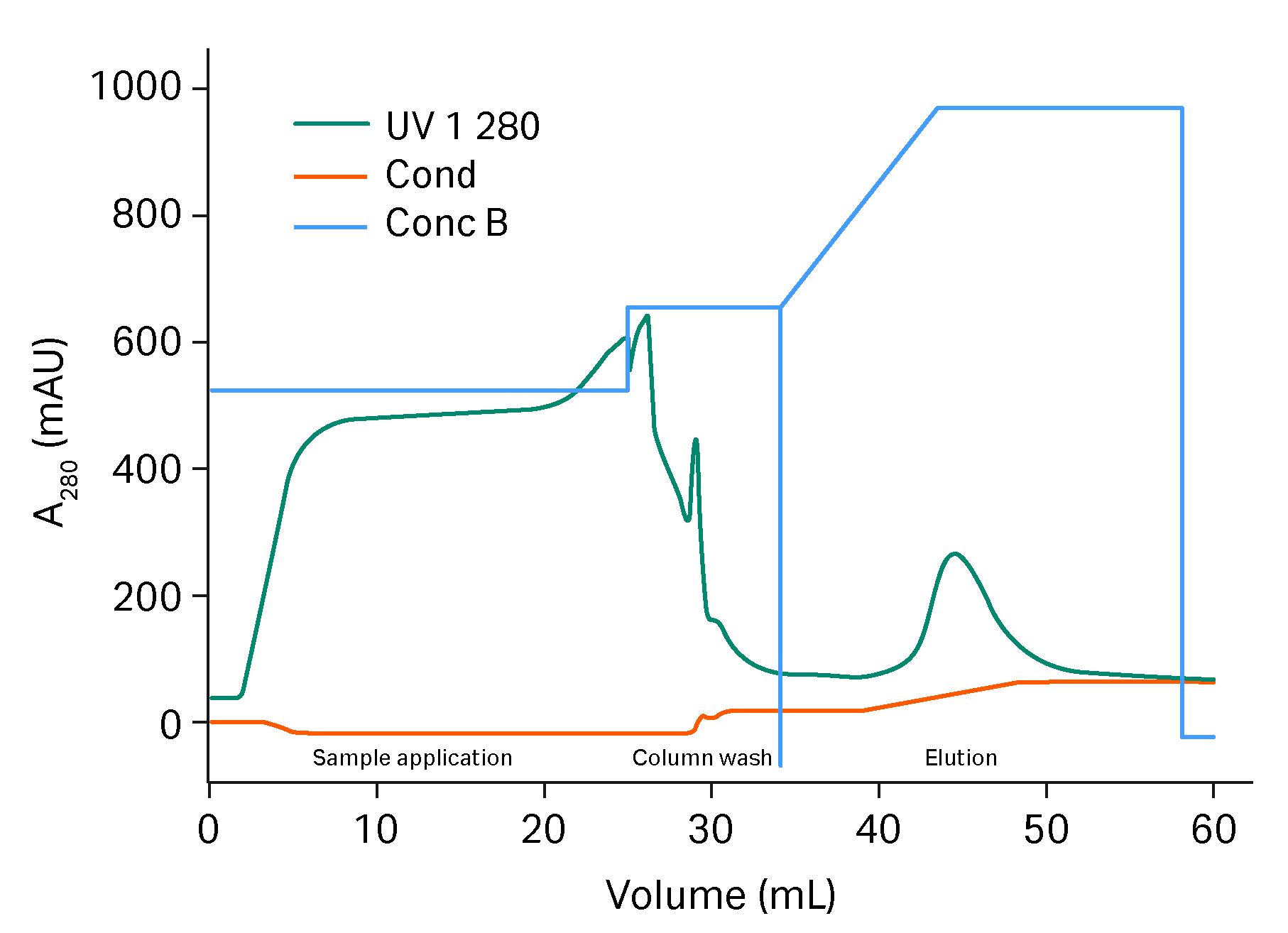
Fig 3. Chromatogram of capture of AdV on HiScreen™ Capto™ Q ImpRes column.
Sample: eluate from HiScreen™ Capto™ Q Impres column capture
Column: HiTrap HiTrap™ Capto™ Core 700 1 mL column
Load: 40 mL
Flow rates: sample load 0.33 mL/min, CIP 0.5 mL/min
Buffer: 20 mM Tris, pH 8.0 + 500 mM NaCl
CIP: 1 M NaOH+ 27% 1-propanol, 5 CV with 30 min pause + 5 CV
System: ÄKTA pure™ 25
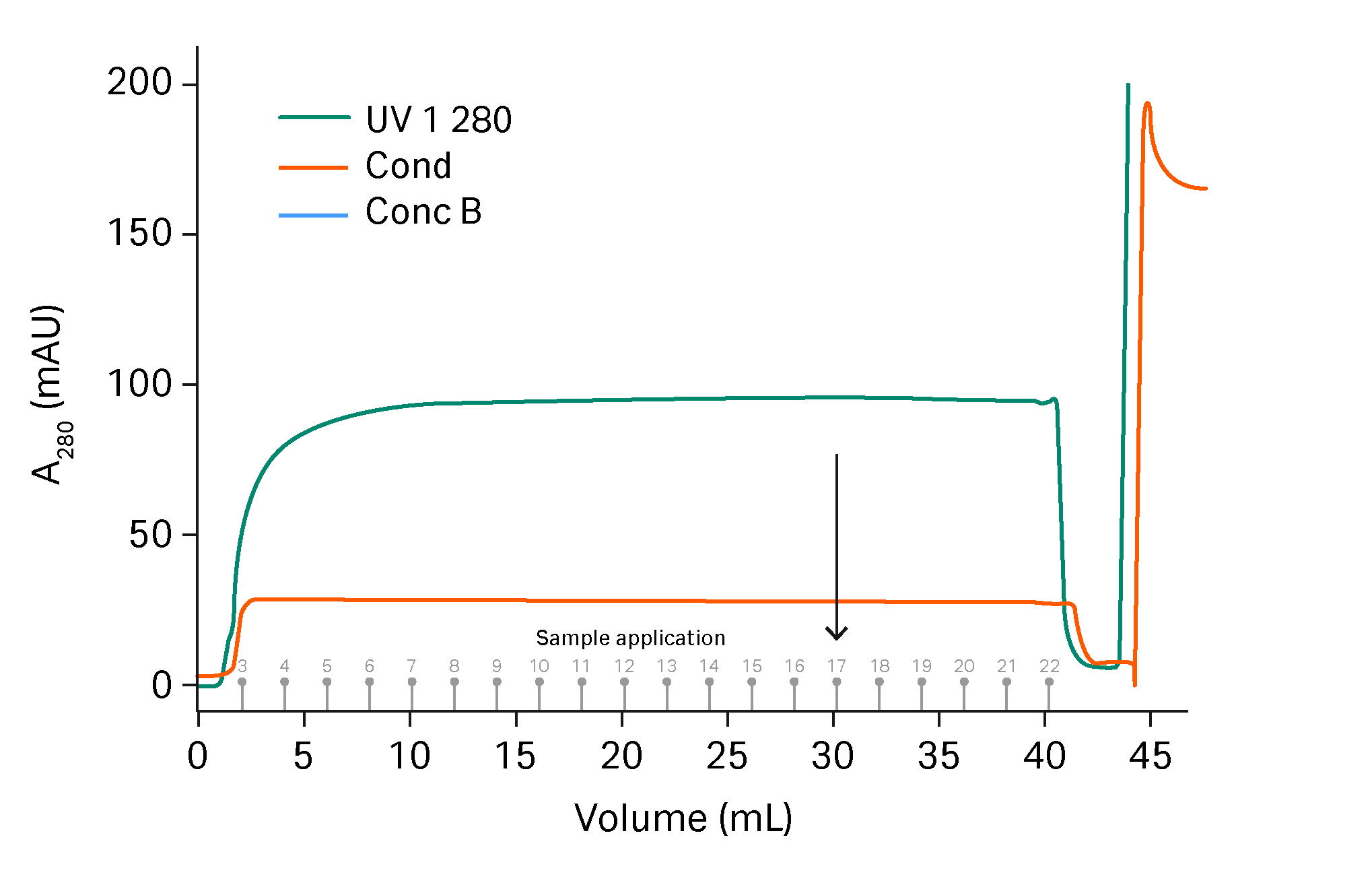
Fig 4. Sample application of HiTrap™ Capto™ Q ImpRes eluate on HiTrap™ Capto™ Core 700 column. Arrow indicates where breakthrough of genomic DNA could be detected in the fractions. The CIP step is not shown.
Conclusions
The developed two-step purification of AdV achieved high resolution between virus and impurities resulting in impurity levels below LOD and high amount of AdV. The two-step method is suitable for scale-up purifications (4,5).
Learn more about Capto™ Core beads and production of virus:
References
- Kallel, H. and Kamen, A.A. Large-scale adenovirus and poxvirus-vectored vaccine manufacturing to enable clinical trials. Biotechnol J 10, 741–747 (2015).
- Adenoviral vector vaccine platforms in the SARS-CoV-2 pandemic; Samir Andrade Mendonça1, Reka Lorincz1, Paul Boucher1 and David T. Curiel 1; npj Vaccines (2021) 97
- Appaiahgari, M.B. and Vrati, S. Adenoviruses as gene/vaccine delivery vectors: promises and pitfalls. Expert Opin Biol Ther 15, 337–351 (2015).
- Application note: Scalable process for adenovirus production. GE Healthcare, KA877080618AN (2018)
- Application note: Optimization of midstream cell lysis and virus filtration steps in an adenovirus purification process. GE Healthcare, KA875220218AN (2018).