Explore the mRNA universe with Cytiva. In this series, our scientists share their journey and insights on manufacturing mRNA for research purposes. And they share their advice on how to scale. Through peer-to-peer interactions, we continue to encourage the spirit of collaboration developed at a time when new modalities are being used as therapies for the first time.
Part 3: mRNA product – IVT
“We’ve found that there isn’t a universal generic protocol that works for every IVT reaction—each new mRNA sequence requires optimization. The same is true for reagents from different vendors. Optimizing the reaction so frequently may seem tedious, but we’ve found it’s worth doing, because it saves you time later” Susanna Lindman, Principal Scientist at Cytiva R&D, Uppsala, Sweden
In vitro transcription (IVT) is the synthesis of RNA molecules, allowing a researcher to tailor synthesis and introduce modifications to produce a transcript. It’s worthwhile to spend some time optimizing mRNA template design, IVT, and other process parameters to simplify downstream processing and ensure the appropriate balance of yield and secondary structure formation.
During IVT, DNA template, RNA polymerase, NTPs, RNase inhibitor, pyrophosphatase, and IVT buffer are combined to produce mRNA. RNase inhibitor prevents degradation of newly formed mRNA. As nucleotide triphosphates (NTPs) are incorporated into the nascent mRNA, pyrophosphates – which bind to free magnesium ions – are released, inhibiting RNA polymerization. Pyrophosphatases must be added to catalyze the hydrolysis of pyrophosphates into inactive single orthophosphate ions that don’t bind magnesium ions, which increases the yield of mRNA. T7, T3, or SP6 prokaryotic phage RNA polymerases are widely used.
Depending on the purification step downstream, you may or may not need to remove the DNA template by adding DNase I after IVT. This step is required, for example, when using a purification method that doesn’t separate mRNA from DNA. It’s also necessary when there’s a concern that DNA fragments may hybridize to mRNA.
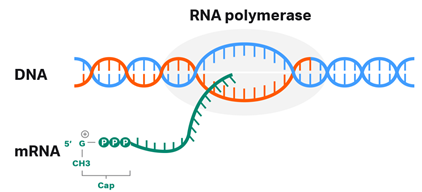
There are two methods to add a 5’ cap (Fig. 3) in vitro – either via a multi-step enzymatic reaction or via co-transcription. The aim is to produce a methylated first nucleotide at the ribose 2’-O position (Cap 1), which has superior in vivo properties compared to a 7-methylguanylate cap structure (Cap 0) at the 5’ end. This is true because the Cap 0 structure can still be recognized as non-self, inducing interferon response and hence a more rapid degradation.
Fig 1. 5’ capping of an mRNA molecule.
In co-transcriptional capping, a cap analog is added directly into the same IVT reaction mixture along with NTPs. This analog is incorporated as the first nucleotide of the transcript, resulting in Cap 1 mRNAs. A large proportion of mRNAs are initially capped. But the final reaction contains a mix of capped and uncapped variants because guanosine triphosphates (GTPs) are depleted.
In contrast, enzymatic capping is performed separately after IVT using guanylyltransferase, followed by 2'-O-methyltransferase. The vaccinia virus capping enzyme executes the three steps needed for the addition of Cap 0 to the 5’ end. These steps are mediated by enzymes RNA triphosphatase, guanylyltransferase, and guanine methyltransferase.
The steps lead to the conversion of the 5’ triphosphate of nascent RNA to the cap 0 structure. First, a phosphate is removed from the 5’ triphosphate to generate 5’ diphosphate mRNA. Then, a guanosine monophosphate (GMP) group is transferred from GTP to the 5’ diphosphate. Finally, a methyl group is added to the N7 amine of the guanine cap to form the cap 0 structure. Cap 0 mRNAs are subsequently converted to Cap 1 mRNAs by 2´-O-methyltransferase by methylating the 2’-OH of the first nucleotide adjacent to the cap structure. The methyl groups are donated by S-adenosylmethionine.
Although the co-transcriptional method appears to be simpler with fewer steps and enzymes, the yields of capped mRNA are usually lower than the more cost-efficient, reliable, and effective enzymatic method. Another drawback of the co-transcriptional method is that a manufacturing license may be required. On the other hand, enzymatic capping can require more time because mRNA needs to be purified before and after capping in some cases, which may also cause some loss of mRNA.
If the polyA tail is not designed in the pDNA or PCR template, a polyA polymerase can be used to catalyze the addition of adenosine monophosphate (AMP) from adenosine triphosphate (ATP) to the 3' ends. The length of the polyA tail can range from 40 to 250 residues long. The optimal polyA tail length may vary by application. This method may generate a heterogeneous population of polyA-tailed mRNAs.
At this point, unpurified (crude) mRNA can be analyzed in several ways. Agarose gel electrophoresis can check for any potential degradation of mRNA, while concentration can be determined by fluorescence-based, RNA-specific assays.
Challenges
- Need for high quality of enzymes, reagents, and filters
- Must choose the capping strategy, bearing in mind costs, time, and reliability
Strategies
- Use high quality IVT enzymes from Aldevron, which can be custom made to meet your specific requirements from mg to g scale.
- Transcription: T7 RNA polymerase, pyrophosphatase, RNase inhibitor
- Template removal: DNase I
- Multi-step capping: guanylyltransferase (3 enzymatic activities) and 2'-O-methyltransferase
- Tailing: polyA polymerase
- Use NTPs from Cytiva, which are tested for RNase activity and are extensively purified (≥ 98 %). They are available either as individual products or in a convenient set containing ATP, CTP, GTP, and UTP in separate tubes.