For life sciences researchers, every new discovery gives rise to more questions and lines of potential research. Every new technique finds more and more applications, opening more avenues of exploration, and expanding our knowledge and understanding.
Single-cell sequencing is an example of a technique that is powering an explosion of scientific research and discovery. Single-cell sequencing enables researchers to distinguish every cell among many types and provide far greater depth of understanding about cell-to-cell relationships. Single-cell sequencing studies will drive an increased understanding of the physiological processes and pathological mechanisms of disease at the single-cell level. These studies might result in the discovery of new diagnostic markers or therapeutic targets.
Single-cell sequencing enables new lines of inquiry
Conventional sequencing approaches utilize many cells, and lack the sensitivity to analyze individual or a small number of cells. As a result, these sequencing modalities lose the heterogeneity information across the total population. In contrast, researchers utilizing single-cell sequencing can characterize individual cellular heterogeneity and can distinguish a small sub-population of cells within a larger overall population.
Single-cell sequencing is revolutionizing oncology
Single-cell analysis of the genomes, transcriptomes, and proteomes provides researchers with a detailed molecular map of the cell types within a tumor and can reveal the mechanisms underlying cancer development, metastasis, and drug resistance. As cancers develop, genomic instabilities increase the genetic and epigenetic diversity within tumors. Subclones within these masses display different molecular signatures, creating heterogeneity that drives metastasis and enables tumors to acquire chemoresistance. Single-cell sequencing gives researchers the ability to detect and characterize the diverse cells within a tumor, where the averaged analysis of whole tissue cannot describe this diversity and ignores rare but clinically important populations.
Multiomics approach harnesses big data to accelerate discovery
The power of single-cell sequencing is even greater when utilized with multiomics. In multiomics, researchers integrate data and information from multiple omic studies, such as genomics, transcriptomics and proteomics, to analyze and understand biological interactions and functions at a molecular level.
Multiomics provides even more information to the researcher, particularly in regard to the role of the tumor microenvironment in disease progression, mechanisms of drug resistance, or the process of cell differentiation. Researchers can begin to unravel the complexities of cancer genesis and progression, understanding the relationships and interactions between genetic mutations and biomarkers, and how mutated cells interact with microenvironment, Scientists use big-data tools to reveal relationships and associations across multiple ”omes.”
The single-cell sequencing workflow
The high cost and technical challenges of single-cell sequencing initially restricted single-cell sequencing to just a few laboratories. Today, single-cell genome and transcriptome sequencing is more robust and broadly accessible throughout the research community.
No matter what the method, the starting material must be of the highest possible quality. DNA and RNA from fresh tissue samples has been subject to less processing stress than nucleic acids extracted from frozen tissue or formalin-fixed paraffin embedded (FFPE) tissues, and therefore tend to be of higher quality. The tissue samples must first be disaggregated into viable single cells. Once a suspension of viable single cells is obtained, the researcher has many technology options for nucleic acid isolation, including column-based or magnetic bead-based extraction. DNA or RNA extraction is followed by amplification of the individual cells and subsequent detection or sequencing. Disaggregation of the tissue sample into single cells, however, is still a weak link in the workflow.
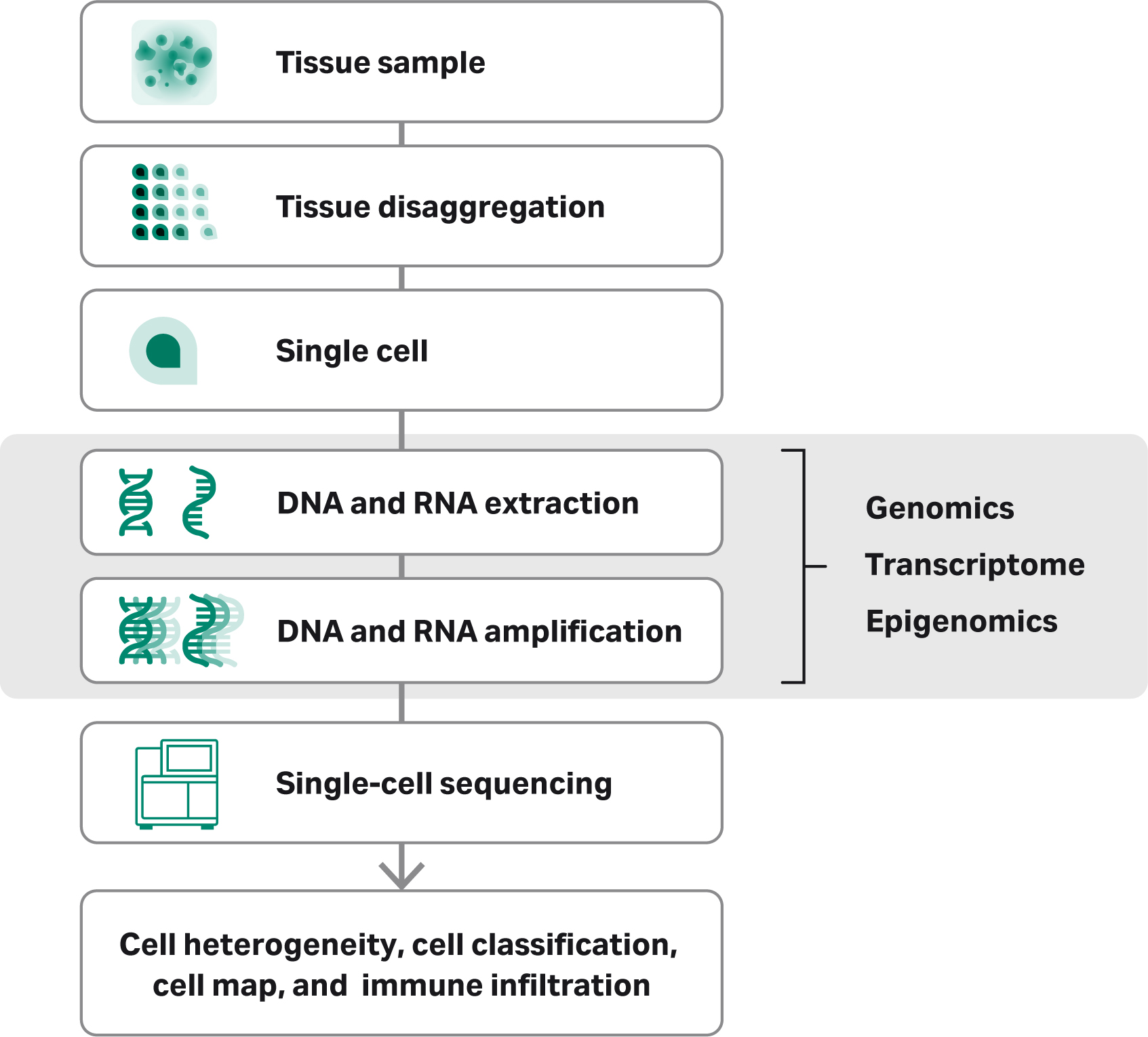
Overview of the single-cell sequencing workflow, including collection, isolation, amplification sequencing and data analysis.
Challenges and considerations for tissue disaggregation
The disaggretation, or dissociation, of the tissue sample into viable single cells is a critical step in single-cell sequencing. Manual tissue processing is time consuming and highly variable dependent on the skills of the operator. Ideally, a mild disaggregation process would ensure that cell yield and integrity remain high and maintain the context of the parent sample. There are several key aspects to consider for an optimal disaggregation process:
- Minimal time between tissue collection and disaggregation into single cells for downstream application(s).
- Protocol consistency to minimize variability throughout the entire workflow.
- High yield and viability of the disaggregated single cells.
- Representative and consistent cell yield and viability cell relative to the original tissue sample.
Transitioning the disaggregation step from a manual process to an automated dissociation system would be highly desirable to researchers, providing the level of consistency so critical to research.
Looking ahead...
New technologies continue to put powerful new tools into the hands of researchers, powering ever-deeper understanding of cells at a molecular level. As our understanding expands, so do the opportunities for clinical applications.
Our white paper “Single cell sequencing: challenges and opportunities” provides an overview of the technologies currently available to researchers as well as current challenges facing researchers and the opportunities presented by the new capabilities.