By Rachel Raybould, Development Scientist Single Cell
Introduction
Single-cell omics has rapidly become the method of choice to investigate cellular heterogeneity among cell populations. To prevent bias in any experiment for single-cell analysis, you need to extract, handle, and process the tissue quickly and with care. So, creating a single-cell suspension from tissue can be long and laborious. To save time, you want to choose equipment that speeds up the single-cell workflow while being gentle on the cells and preserving the original cell state as much as possible. The VIA Extractor™ tissue disaggregator (Cytiva) provides low-impact, fast, and gentle tissue dissociation into single-cell suspensions.
In a previous VIA Extractor™ tissue disaggregator case study for mouse liver tissue, we demonstrated that the VIA Extractor™ tissue disaggregator produces cells of a higher yield and less cellular fragility when compared with liver cell suspensions dissociated on the gentleMACS system (Miltenyi Biotec). In this case study, we investigate the ability of the VIA Extractor™ tissue disaggregator to generate suspensions of high quality single cells from a tougher tissue type: the heart. We compared the performance, in terms of yield, cell viability and single-cell RNA (scRNA) sequencing data, of the VIA Extractor™ tissue disaggregator with the gentleMACS Octo (Miltenyi Biotec).
Method
Mouse heart tissue was obtained from 12 female Crl:CD1 (ICR) mice and washed in ice cold phosphate buffer saline (PBS). Forceps were used to pump blood out of each heart and any connective tissue was removed. The mouse hearts were halved into paired samples: half were used for dissociation on VIA Extractor™ tissue disaggregator and half for dissociation on gentleMACS Octo. Half hearts from mice 1, 2, 3, and 4 made up samples V1 and GM1. Half hearts from mice 5, 6, 7, and 8 made up samples V2 and GM2. Half hearts from mouse 9, 10, 11, and 12 made up samples V3 and GM3. Equal amounts of tissue and Miltenyi Biotec multi tissue dissociation kit reagents (Miltenyi Biotec) were added to each experimental replicate (Fig 1 and Table 1).
For tissue dissociation on the gentleMACS Octo, Miltenyi Biotec’s multi-tissue dissociation kit 2 protocol was followed. For tissue dissociation on the VIA Extractor™ tissue disaggregator, the tissue was dissociated at 37°C for 30 minutes at 200 rpm using Miltenyi Biotec’s multi tissue dissociation kit 2 enzyme kit (Table 2).
Following dissociation, the sample IDs were re-labelled with the intent to process the samples through red blood cell lysis, debris removal, cell capture, and library preparation blind to minimize bias. All cell suspensions were passed through 100 µm cell strainers and subjected to red blood cell lysis and debris removal using Miltenyi Biotec’s red blood cell lysis and debris removal kits. Cells were counted in duplicate using a NucleoCounter® NC-200 (ChemoMetec) and Via2-Cassettes (ChemoMetec). The manufacturer’s instructions for the Chromium Next GEM Single Cell 3’ dual index kit v3.1 (10X Genomics) were followed with the aim to sequence 1000 cells for each sample. A Chromium Controller (10X Genomics) was used to capture the cells into gel beads in emulsion (GEMs).
Libraries were sequenced on a NextSeq 550 Base (Illumina, Inc.) using NextSeq 550 high output kit v2.5 (Illumina, Inc.). The scRNA matrix data were analysed using Uniform Manifold Approximation and Projection (UMAP) (1) in Seurat (2). Each sample was analyzed individually, filtered to remove duplicates, include cells with feature RNAs between 200 and 6000, and remove all cells with a mitochondrial gene expression percentage greater than 5%. Once all samples were filtered and clustered, the data from each sample were combined into a single dataset to allow comparison of the scRNA data from the VIA Extractor™ tissue disaggregator and gentleMACS Octo using Seurat (2) and UMAP (1). Cell types representing each cluster were identified using Seurat (2) and marker genes identified by Litviňuková et al. (3). Genes lists from cell clusters with differential gene expression profiles were further analyzed using the gene ontology software package PANTHER (4). All ANOVA tests were performed in JMP Statistical Discovery software by SAS.
Table 1.Tissue weights for each sample dissociated on the VIA Extractor™ tissue disaggregator and gentleMACS Octo. Hearts from mice 1 to 4 were halved and divided equally between samples V1 and GM1. Hearts from mice 5 to 8 were halved and divided equally between samples V2 and GM2. Hearts from mice 9 to 12 were halved and divided equally between samples V3 and GM3.
| Mouse |
Weight of hearts for VIA Extractor™ tissue disaggregator (g) |
Weight of hearts for gentleMACS Octo (g) |
Enzyme Used | Vol of Enzyme (mL) |
| 1, 2, 3, and 4 | Sample V1=0.517 | Sample GM1=0.511 | Multi Tissue Kit 2 (Miltenyi Biotec) | 2.5 |
| 5, 6, 7, and 8 | Sample V2=0.331 | Sample GM2=0.389 | Multi Tissue Kit 2 (Miltenyi Biotec) | 2.5 |
| 9, 10, 11, and 12 | Sample V3=0.409 | Sample GM3=0.426 | Multi Tissue Kit 2 (Miltenyi Biotec) | 2.5 |
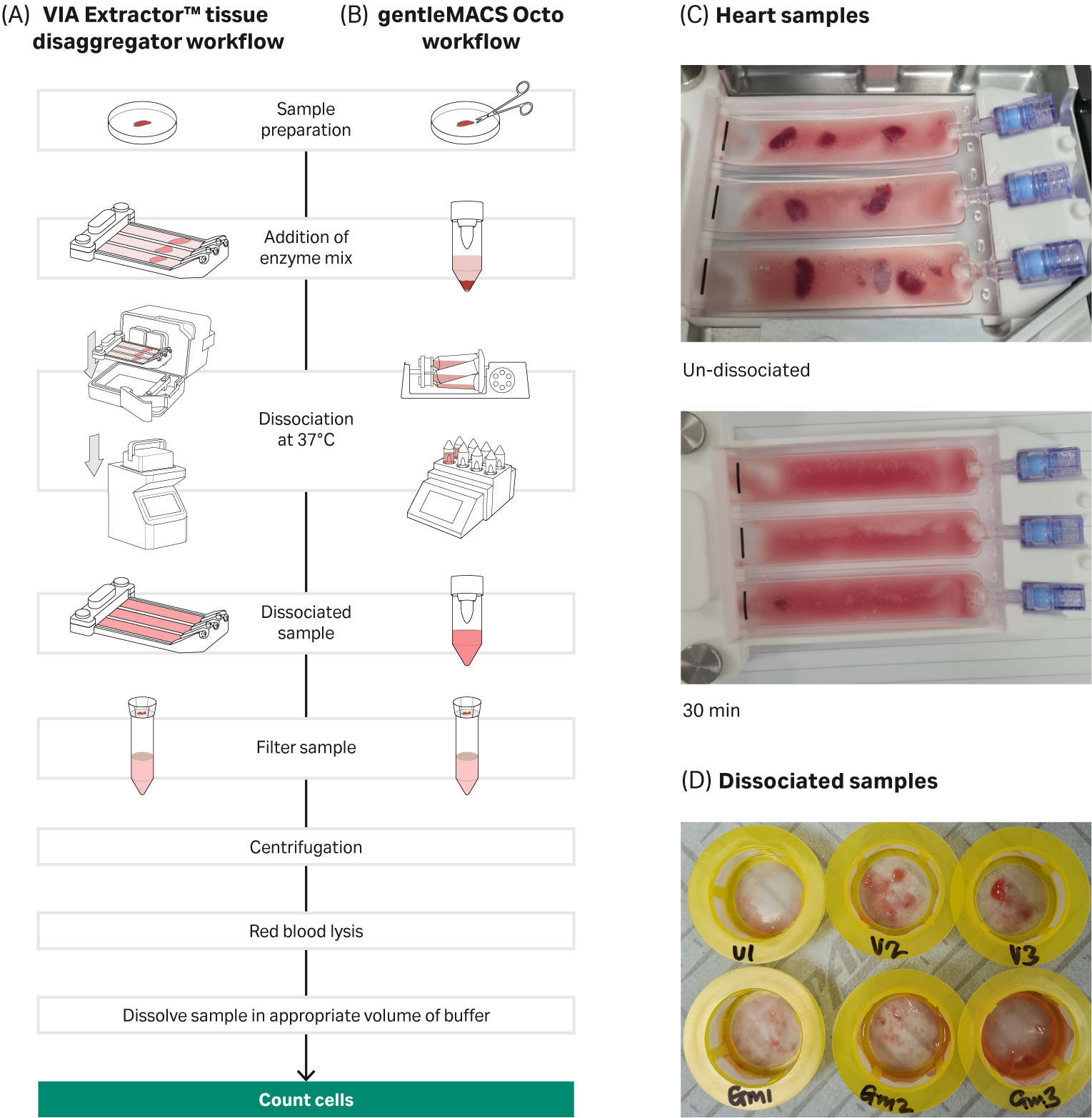
Fig 1. Tissue dissociation workflow for VIA Extractor™ tissue disaggregator and gentleMACS Octo: A) VIA Extractor™ tissue disaggregator workflow B) gentleMACS Octo workflow C) Heart samples were fully dissociated on the VIA Extractor™ tissue disaggregator in 30 minutes. D) Top view of cell strainer with dissociated samples from the VIA Extractor™ tissue disaggregator (samples V1, V2, and V3) and gentleMACS Octo (samples GM1, GM2, and GM3).
Table 2. Temperature, speed, and time conditions for dissociation on VIA Extractor™ tissue disaggregator and gentleMACS Octo.
|
VIA Extractor™ tissue disaggregator |
gentleMACS Octo | |
| Program used | N/A | 37C_Multi_G |
| Program speed | 200 rpm | N/A |
| Program time | 30 minutes | 56 minutes |
| Program temperature | 37°C | 37°C |
Results
This study demonstrates that the VIA Extractor™ tissue disaggregator offers a fast and gentle approach to tissue dissociation with improved scRNA sequence data quality but without any impact on cell viability.
The sample preparation time, in terms of washing the tissue and removing blood, for both methods of dissociation was comparable. However, tissue dissociation on the VIA Extractor™ tissue disaggregator was significantly faster compared with the gentleMACS Octo. The heart tissue sample was fully dissociated in 56 minutes using the gentleMACS Octo, but only took 30 minutes to dissociate using the VIA Extractor™ tissue disaggregator (Fig 1B).
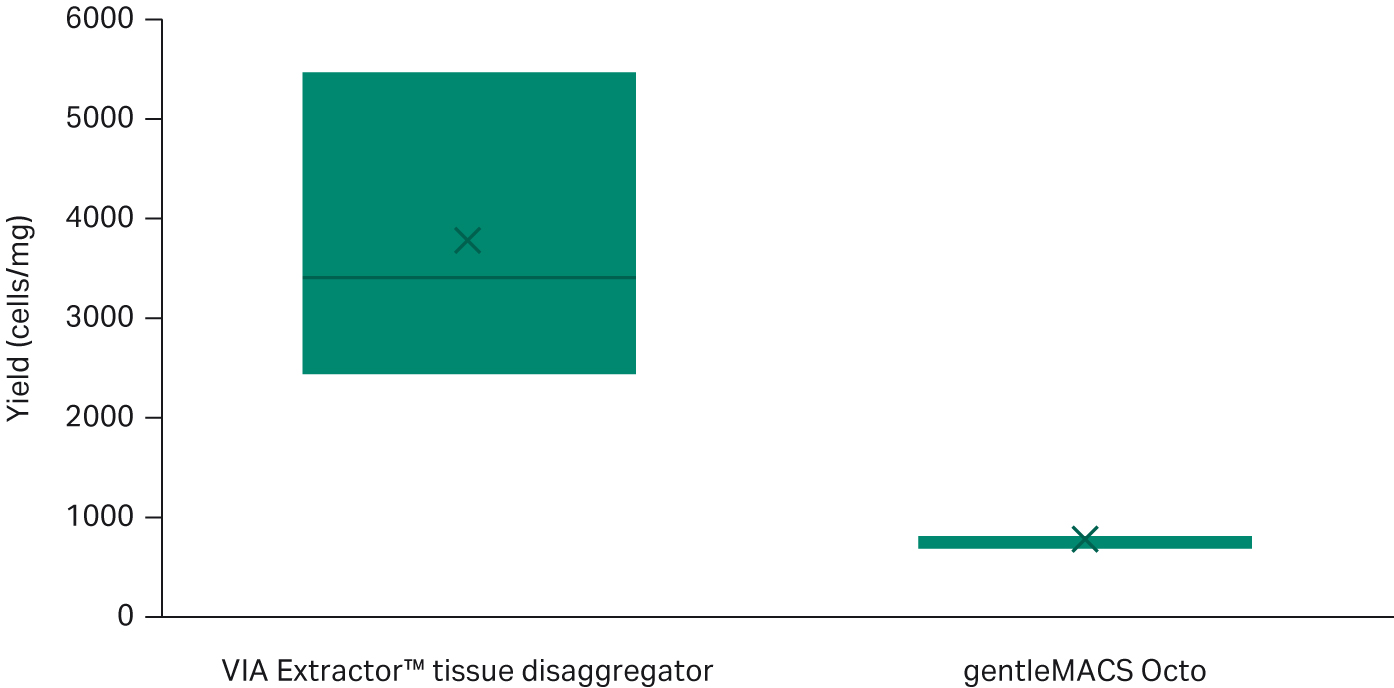
The reduction in dissociation time did not impact the yield of cells dissociated from the heart tissue. On the contrary, the cell yield from the VIA Extractor™ tissue disaggregator was more than double that of the gentleMACS Octo. The VIA Extractor™ tissue disaggregator resulted in a higher cell yield (cells per milligram tissue) compared to the gentleMACS Octo (t test p = 0.0392, df = 2) (Fig 2).
Both methods of dissociation produced cells that were more than 90% viable after red blood cell lysis and debris removal. The cells were of excellent quality for single-cell sequencing.
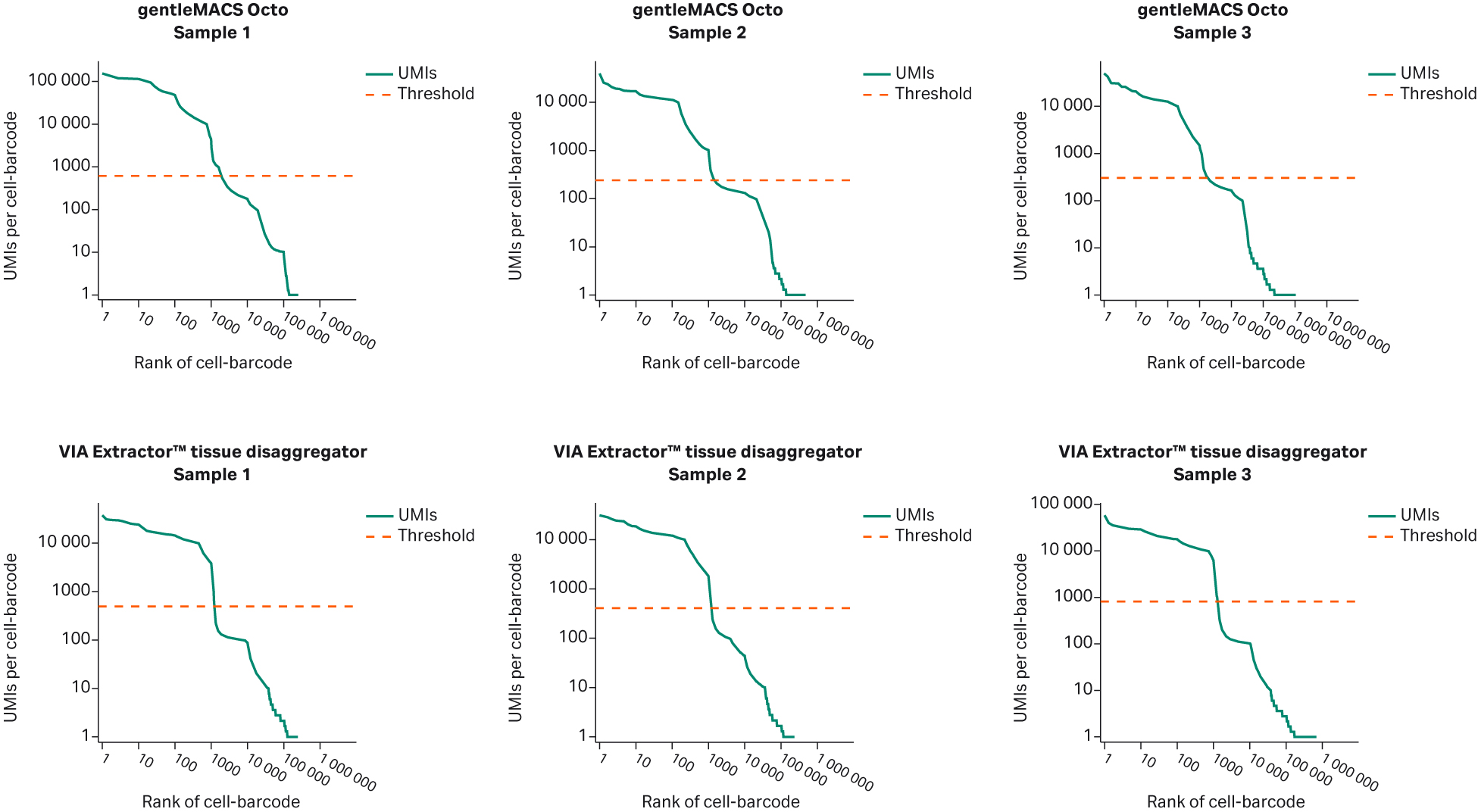
DRAGEN (Illumina, Inc.) single-cell data analysis indicated that the DRAGEN QC metrics were different between cells dissociated on the VIA Extractor™ tissue disaggregator and gentleMACS Octo. Knee plots from the VIA Extractor™ tissue disaggregator were also observed to be steeper than those of the gentleMACS Octo (Fig 3). Typically, a knee plot from a good quality sample with healthy cell membranes will display a flat plateau followed by a steep drop off. A steep drop off means that there is a clear difference between barcodes that are associated with cells and barcodes that are noncellular. A curve indicates that there is “noise” of noncell associated barcodes that is normally associated with extracellular RNA contamination. The knee plot data support the notion that VIA Extractor™ tissue disaggregator offers a more gentle approach to tissue dissociation. It is possible that the gentleMACS Octo produces cells that are viable but potentially too fragile to survive the GEM capture process on the Chromium Controller. The fragile cells may break and lead to extracellular RNA that reduces the distinction between cell-based barcodes and non cell-based barcodes.
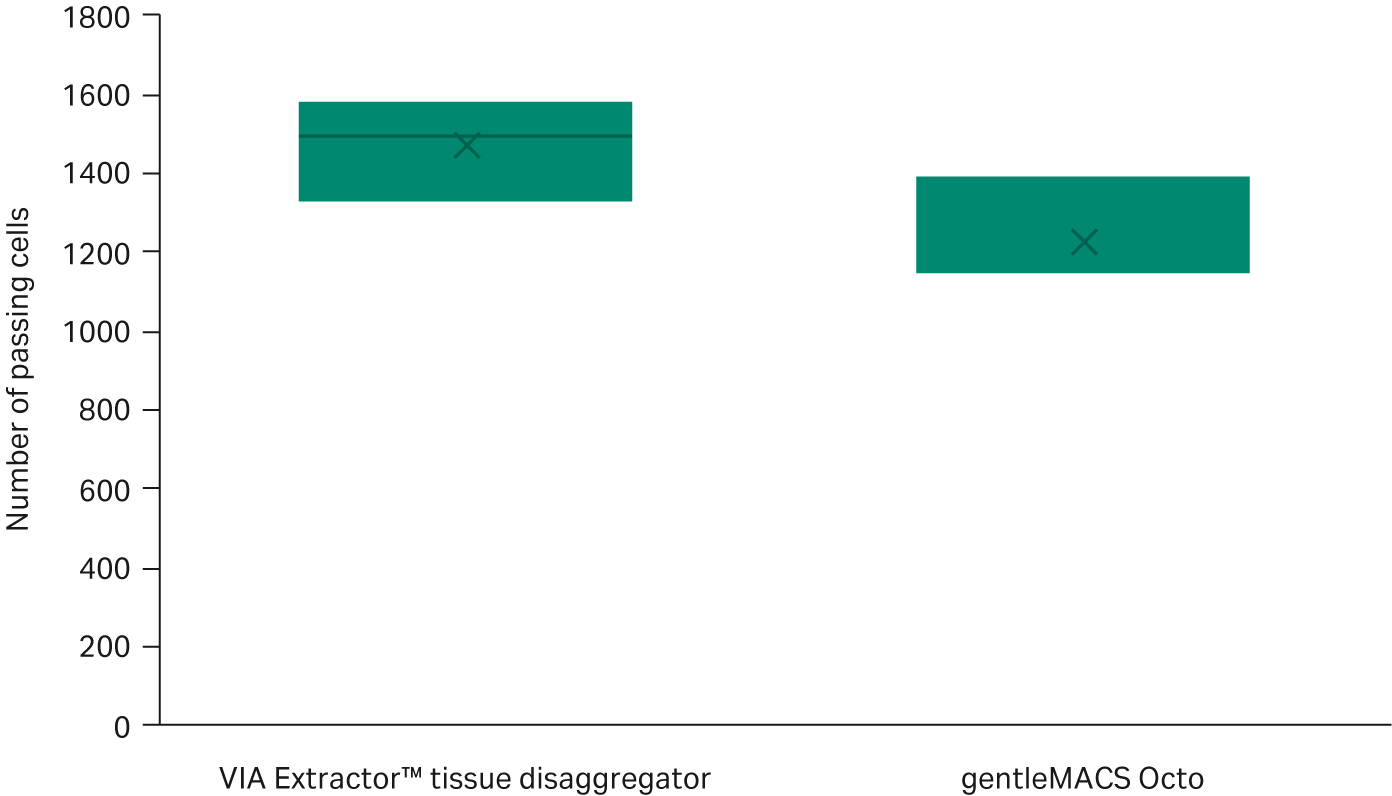
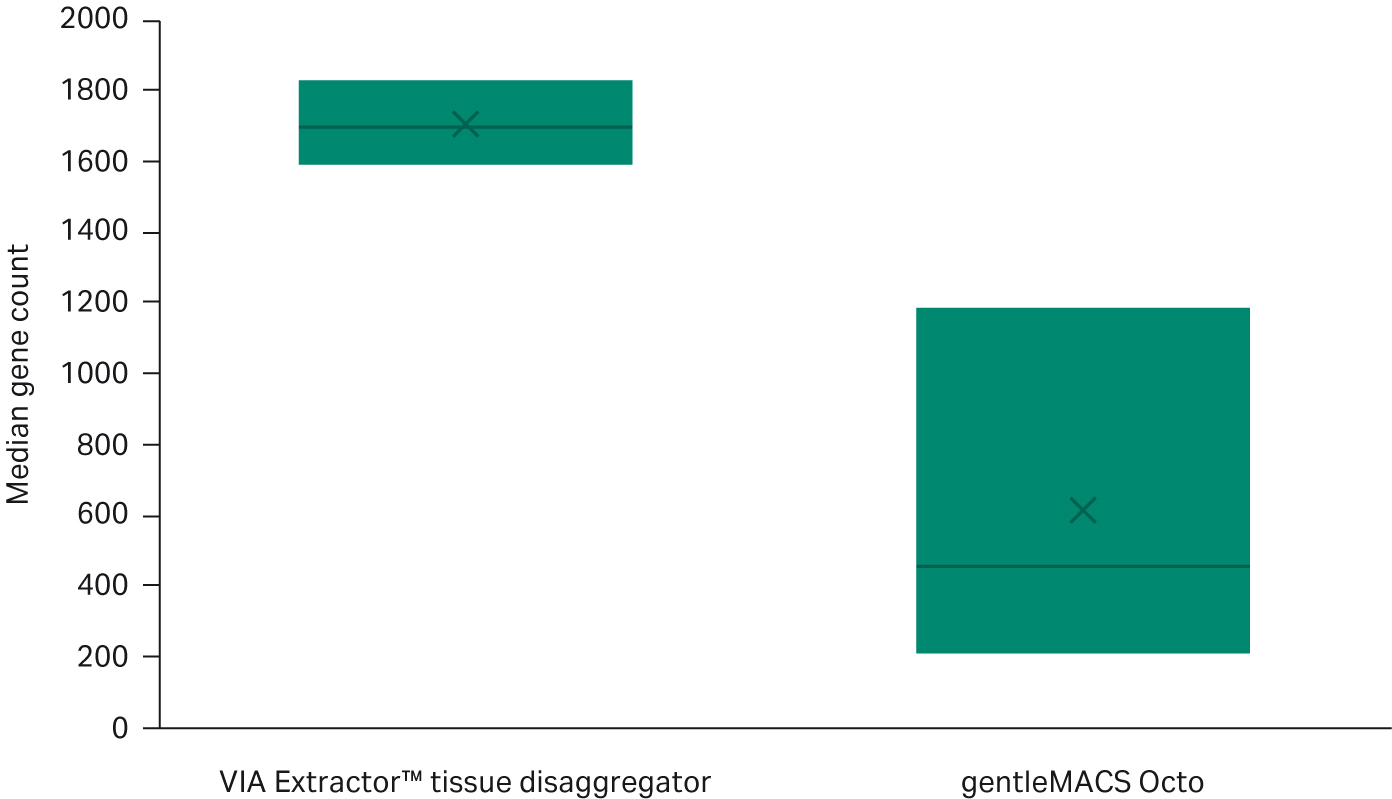
The data from the knee plots are supported by other DRAGEN single-cell metrics such as median gene counts per cell and number of cells passing (Fig 4 and 5). The VIA Extractor™ tissue disaggregator had a higher number of cells passing metric thresholds compared with gentleMACS Octo (Fig 4). The median number of gene counts per cell for VIA Extractor™ tissue disaggregator was 1705 ± 122 compared with 617 ± 509 for the gentleMACS Octo. This is statistically significant (t test p = 0.023, df = 1) (Fig 5). The large standard deviation for the gentleMACS Octo samples indicates that the number of gene counts varies considerably between cells, which could be a result of the contaminating barcodes in the cells. These data support the observation made above for the knee plots. In summary, a higher median gene count with a smaller standard deviation and more cells passing are achieved with the VIA Extractor™ tissue disaggregator, which indicates improved quality of cells.
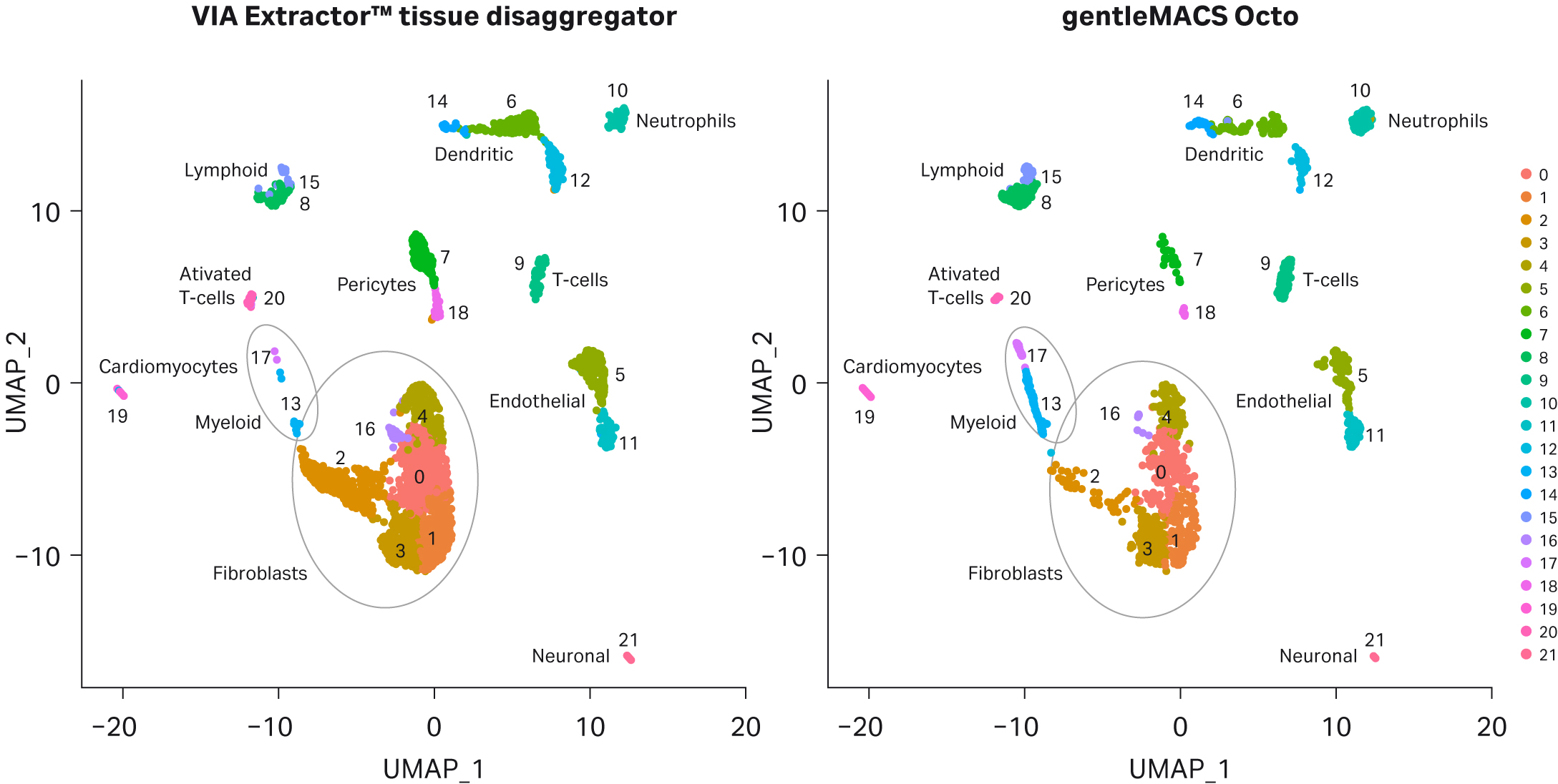
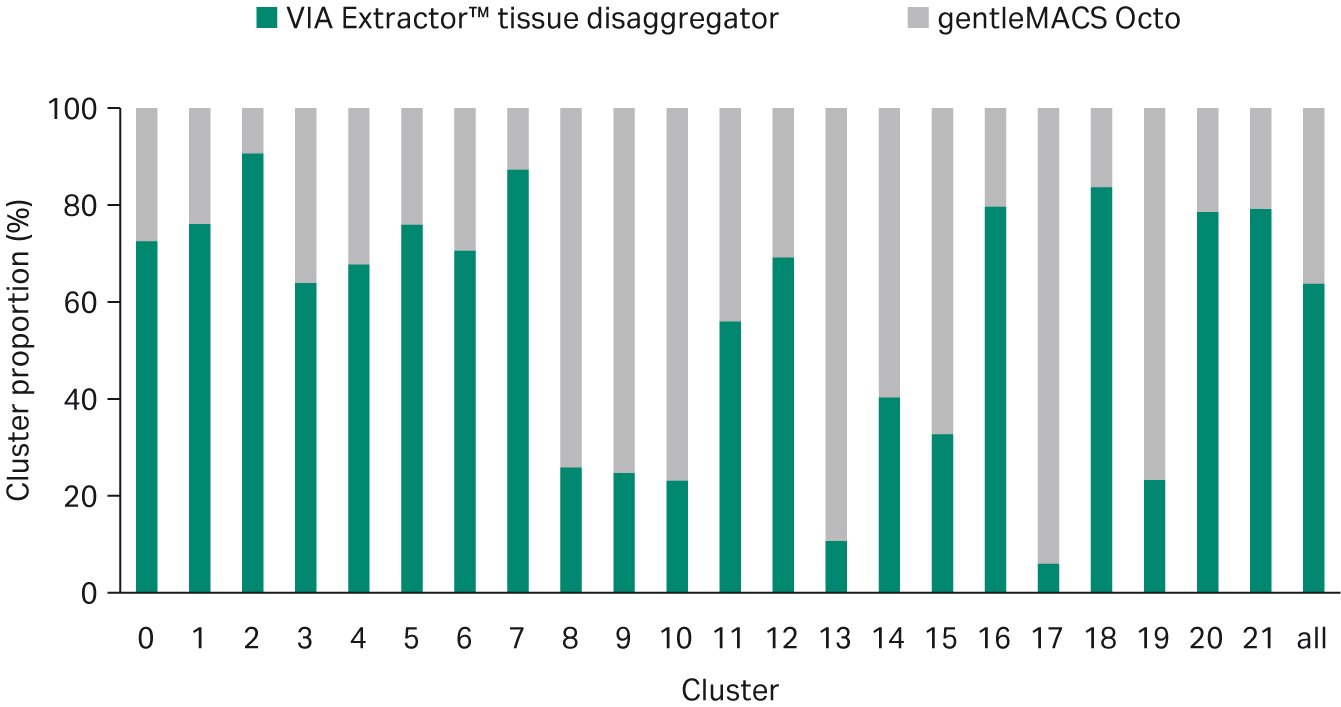
UMAP analysis uncovered 21 cell clusters and cell types for tissue samples dissociated on both the gentleMACS Octo and VIA Extractor™ tissue disaggregator (Fig 6). There were differences in the proportion of certain cell clusters (Fig 7). At the extremes (cell count proportions that differed by over 80% or under 20%) were cell clusters 2, 7, 18, 13, and 17. Clusters 2, 17, and 18 were overrepresented in samples dissociated by the VIA Extractor™ tissue disaggregator. Clusters 7 and 18 were easily identified as pericytes as they were expressing pericyte gene markers ATP Binding Cassette Subfamily C Member 9 (ABCC9), Potassium Inwardly Rectifying Channel Subfamily J Member 8 (KCNJ8), and Regulator Of G Protein Signalling 5 (RGS5), as cited by Litviňuková et al. (3). Cluster 2 was identified as expressing fibroblast markers such as Decorin (DCN), Gelsolin (GSN), and Platelet Derived Growth Factor Receptor Alpha (PDGFRA). Also, cluster 2 had a significantly lower-fold change expression of genes associated with cell stress response (Table 4). The data for cluster 2 supports that heart fibroblasts are less stressed by dissociation on the VIA Extractor™ tissue disaggregator.
Clusters 13 and 17 express myeloid genes such as Transmembrane Immune Signalling Adaptor (TYROBP) and Triggering Receptor Expressed On Myeloid Cells Like 1 (TREML1) and were over represented in samples dissociated on the gentleMACS Octo. Clusters 13 and 17 are characterised by over expressing genes that encode proteins involved in the inflammatory response and wound healing, respectively (Table 4). CQ1 is involved in complement activation, response to infection, and removal of apoptotic cells. The gene list for cluster 17 indicates that there is a high level of expression of genes encoding proteins involved in wound healing. The data for cluster 13 and 17 can be interpreted in one of two ways. The first interpretation supports the concept that the VIA Extractor™ tissue disaggregator provides a method of gentle dissociation with less damage to cells, which results in fewer cells expressing genes involved in macrophage activation and response to tissue damage. The second interpretation may indicate that the gentleMACS Octo allows detection of the myeloid cells involved in complement activation and tissue repair.
Fig 2. Comparison of cell yield between the VIA Extractor™ tissue disaggregator and the gentleMACS Octo (t test p = 0.0392, df = 2). The VIA Extractor™ tissue disaggregator results in higher cell yield compared to gentleMACS Octo.
Table 3. The cell count per milligram of tissue is higher in hearts dissociated with the VIA Extractor™ tissue disaggregator compared with the gentleMACS Octo. There is no difference in viability between VIA Extractor™ tissue disaggregator and gentleMACS Octo.
| New Sample ID |
Dissociation method |
Viabilty (%) |
Yield (cells/mg tissue) |
| 1A | gentleMACS Octo (GM1) | 95 | 695 |
| 2A | VIA Extractor™ tissue disaggregator (V1) | 97 | 3404 |
| 3A | VIA Extractor™ tissue disaggregator (V2) | 95 | 5468 |
| 4A | gentleMACS Octo (GM2) | 94 | 838 |
| 5A | gentleMACS Octo (GM3) | 91 | 822 |
| 6A | VIA Extractor™ tissue disaggregator (V3) | 96 | 2445 |
Fig 3. Knee plots for each of the samples post analysis with DRAGEN single-cell RNA application. The samples dissociated on the VIA Extractor™ tissue disaggregator display well-defined knee plots, whereas samples dissociated on gentleMACS Octo have a poorly-defined drop off. A steep drop off is indicative of good separation between the cell-associated barcodes and the barcodes associated with empty partitions. A curved drop off indicates contamination of empty partitions and potentially cell-associated barcodes containing cell free RNA.
Fig 4. Comparison of the number of cells passing DRAGEN single-cell RNA quality checks. The number of cells passing DRAGEN single-cell RNA quality checks is higher with cells dissociated on VIA Extractor™ tissue disaggregator compared with gentleMACS Octo (t test p = 0.0492, df = 4).
Fig 5. Comparison of the median gene count between VIA Extractor™ tissue disaggregator with the gentleMACS Octo following DRAGEN single-cell RNA quality checks. The number of cells passing DRAGEN single-cell RNA quality checks is higher with cells dissociated on VIA Extractor™ tissue disaggregator compared with gentleMACS Octo (t test p = 0.0295, df = 2).
Fig 6. UMAP clustering of cell samples. Following sequencing, 21 clusters were automatically generated for both sample sets by Seurat analysis and cell types assigned.
Fig 7.Proportional representation of cells from both sample sets in each of the clusters identified in Figure 6.
Table 4. Gene ontology (GO) biological process analysis using protein analysis through evolutionary relationships (PANTHER) for top 10 genes listed for clusters 2, 13 and 17. Ensembl ID, gene name ID, log fold change in gene expression (Log FC- exp) for the VIA Extractor™ tissue disaggregator and gentleMACS Octo, biological process, GO p-value and GO false discovery rate (FDR) are shown.
| VIA Extractor™ tissue disaggregator | gentleMACS Octo | ||||||
| Cluster | Ensembl ID | Gene ID | Log-FC exp | Log-FC exp | GO biological process of all 10 genes | GO p-value | GO FDR |
| 2 | ENSMUSG00000052837 | Junb | -3.227916943 | -3.764941075 | negative regulation of transcription from RNA polymerase II promoter in response to stress (GO:0097201) | 1.38E-05 | 9.03E-03 |
| ENSMUSG00000021250 | Fos | -3.770762808 | -3.554206217 | response to corticosterone (GO:0051412) | 2.25E-05 | 1.31E-02 | |
| ENSMUSG00000052684 | Jun | -3.140466151 | -3.274124354 | integrated stress response signaling (GO:0140467) | 2.29E-07 | 5.12E-04 | |
| ENSMUSG00000020423 | Btg2 | -3.138476787 | -3.097545505 | response to muscle stretch (GO:0035994) | 4.61E-05 | 1.90E-02 | |
| ENSMUSG00000092341 | Malat1 | -2.120789187 | -1.760016056 | cellular response to calcium ion (GO:0071277) | 8.35E-11 | 1.31E-06 | |
| ENSMUSG00000071076 | Jund | -2.102836728 | -0.982808053 | response to mineralocorticoid (GO:0051385) | 9.22E-05 | 2.63E-02 | |
| ENSMUSG00000091971 | Hspa1a | -3.677973078 | -2.760131714 | cellular response to cadmium ion (GO:0071276) | 1.02E-04 | 2.76E-02 | |
| ENSMUSG00000024190 | Dusp1 | -2.896103086 | -2.236973183 | response to progesterone (GO:0032570) | 1.07E-04 | 2.76E-02 | |
| ENSMUSG00000003545 | Fosb | -3.01166351 | -1.895512893 | positive regulation of miRNA transcription (GO:1902895) | 1.48E-04 | 3.26E-02 | |
| ENSMUSG00000086503 | Xist | -1.762025276 | -0.925347092 | skeletal muscle cell differentiation (GO:0035914) | 1.60E-04 | 3.44E-02 | |
| 13 | ENSMUSG00000036887 | C1qa | 3,488800882 | 4,760969916 | synapse pruning (GO:0098883) | 1.93E-08 | 1.53E-04 |
| ENSMUSG00000036905 | C1qb | 3,628318699 | 4,393600093 | cell junction disassembly (GO:0150146) | 1,21E-04 | 3.07E-08 | |
| ENSMUSG00000036896 | C1qc | 3,345863532 | 4,070158702 | neutrophil activation involved in immune response (GO:0002283) | 8.14E-03 | 1.45E-05 | |
| ENSMUSG00000069516 | Lyz2 | 3,923377675 | 2,807094198 | microglial cell activation (GO:0001774) | 6.79E-04 | 3.02E-07 | |
| ENSMUSG00000058715 | Fcer1g | 3,257880612 | 2,359244407 | leukocyte activation involved in inflammatory response (GO:0002269) | 7.69E-04 | 4.39E-07 | |
| ENSMUSG00000030579 | Tyrobp | 3,115315771 | 2,243030603 | glial cell activation (GO:0061900) | 8.10E-04 | 5.65E-07 | |
| ENSMUSG00000024397 | Aif1 | 3,731604102 | 3,135386677 | positive regulation of protein localization to cell surface (GO:2000010) | 2.43E-02 | 6.00E-05 | |
| ENSMUSG00000025150 | Cbr2 | 3,372520543 | 2,692310302 | neutrophil activation (GO:0042119) | 2.56E-02 | 6.48E-05 | |
| ENSMUSG00000069792 | Wfdc17 | 3,752593608 | 3,369812165 | neuroinflammatory response (GO:0150076) | 8.73E-04 | 8.86E-07 | |
| ENSMUSG00000004814 | Ccl24 | 3,047080867 | 1,309298968 | positive regulation of myeloid leukocyte mediated immunity (GO:0002888) | 3.09E-02 | 8.03E-05 | |
| 17 | ENSMUSG00000036353 | P2ry12 | 1,986764773 | 3,008233495 | platelet activation (GO:0030168) | 2.91E-02 | 1.30E-05 |
| ENSMUSG00000020787 | P2rx1 | 2,066112579 | 1,733719331 | blood coagulation (GO:0007596) | 9.19E-05 | 1.17E-08 | |
| ENSMUSG00000000320 | Alox12 | 3,339230632 | 3,019615597 | coagulation (GO:0050817) | 6.47E-05 | 1.24E-08 | |
| ENSMUSG00000030054 | Gp9 | 2,831003703 | 3,379404413 | hemostasis (GO:0007599) | 5.27E-05 | 1.34E-08 | |
| ENSMUSG00000024511 | Rab27b | 2,868540327 | 2,540107035 | regulation of body fluid levels (GO:0050878) | 1.19E-04 | 7.59E-09 | |
| ENSMUSG00000032261 | Sh3bgrl2 | 2,827713303 | 3,069296517 | wound healing (GO:0042060) | 8.78E-04 | 2.80E-07 | |
| ENSMUSG00000023993 | Treml1 | 3,389123721 | 2,330857247 | response to wounding (GO:0009611) | 2.80E-03 | 1.07E-06 | |
| ENSMUSG00000046814 | Gchfr | 1,944583533 | 2,099188176 | ||||
| ENSMUSG00000073414 | Mpig6b | 2,065347999 | 1,819772799 | ||||
| ENSMUSG00000034664 | Itga2b | 2,751894914 | 3,408245231 | ||||
Conclusion
Semiautomated tissue disaggregation is an effective way of easing the burden of tissue dissociation, which is the first step in a long single-cell sequencing workflow. To reduce biases that may be introduced by sample preparation workflows, it is important to maintain the cell state as near to its original tissue state as possible. To do this, tissue dissociation requires a fast process that will introduce the least amount of stress possible to the cells. We have previously demonstrated with mouse liver tissue that the VIA Extractor™ tissue disaggregator produces cells of a higher yield and less cellular fragility when compared with liver cell suspensions dissociated on the gentleMACS system (Miltenyi Biotec). In this current investigation, we performed tissue dissociation to compare the VIA Extractor™ tissue disaggregator to gentleMACS Octo using three paired samples from mouse heart tissue. Heart tissue is known for its tough and difficult to dissociate nature. We have confirmed that for heart tissue, the VIA Extractor™ tissue disaggregator provides a faster solution with increased yield of cells compared with the gentleMACS Octo. Furthermore, our data demonstrates that the gentle approach of the VIA Extractor™ tissue disaggregator improves the quality of the single-cell RNA sequencing data, reduces the number of cells with fragile cell membranes, and improves cell capture.
This data is based on three independent experiments with the equal number of replicates in each experiment. All samples tested were treated equally (with the number of replicates being the same for all products tested in the comparison) and according to manufacturers’ protocol and recommendations. Data was collected at Cytiva, Maynard Centre, Cardiff, UK (R&D Laboratory) during April to August 2022 and is held at this location.
References
- Becht E, McInnes L, Healy J, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018;37(1):38-44.
- Hao Y, Hao S, Andersen-Nissen E, et al. Integrated Analysis of multimodal single-cell data. Cell. 2021;184(13):3573-3587.
- Litviňuková M, Talavera-López C, Maatz H, et al. Cells of the adult human heart. Nature. 2020;588(7838):466-472.
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. Panther version 14: More genomes, a new panther go-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2018;47(D1):D419-D426.