By Ben Tivey, R&D Scientist
Introduction
Single-cell RNA sequencing (scRNA-seq) allows researchers to delve deeper into the heterogeneity among cellular populations. For example, clinicians can explore the intratumor genomic heterogeneity of rare cancer biopsies. Single-nuclei RNA sequencing (snRNA-seq) is a technique that uses isolated nuclei, rather than whole single cells, to ascertain the genomic landscape of cellular populations within a given tissue sample. Nuclei are often preferred as a starting material for genomic applications because they can be extracted and processed post-cryopreservation. Whole cells are prone to thawing-induced cell death. Previous comparative studies between snRNA-seq and scRNA-seq concluded that, in adult murine kidney, snRNA-seq provides reduced dissociation bias, abolition of dissociation-induced transcriptional stress responses, and even the ability to process inflamed fibrotic tissue (1).
We previously showed that the VIA Extractor™ tissue disaggregator (Cytiva) can process fresh tissue samples into single-cell suspensions of high yield and viability. Here, we investigate the ability of the VIA Extractor™ tissue disaggregator to fully dissociate both fresh and snap-frozen tissue into single-nuclei suspensions for downstream sequencing and subsequent analyses.
Fig 1. (A) The VIA Extractor™ tissue disaggregator provides fast, minimal-impact tissue dissociation into single-nuclei suspensions, (B) The Omics pouch placed into the VIA Extractor™ tissue disaggregator and held in place with the Omics clamp. (C) The VIA Extractor™ tissue disaggregator placed into the top of the VIA Freeze™ Uno controlled-rate freezer.
Methods
Murine kidney and heart tissues were collected from three freshly dissected Crl:CD1 (ICR) female mice (Charles River). Three kidney pairs and six hearts were carefully separated in half. The heart halves were then divided into paired samples to further reduce sample variability. One half of each tissue type was snap-frozen in liquid nitrogen and placed into -80°C storage. The other halves were washed in ice-cold phosphate-buffered saline (PBS) and weighed (Fig 2).
For nuclei extraction, a modified version of the methods set out by Fish et al. (2) was applied for use with the VIA Extractor™ tissue disaggregator.
For the fresh kidney and heart samples, the tissue was placed into the Omics pouch (Cytiva) using the Omics applicator (Cytiva), heat sealed, and placed into the Omics clamp (Cytiva). When repeated on the frozen batches (which were stored at -80°C for one month), each sample was removed from storage, weighed, and 1 mL of tissue lysis solution was added directly onto the tissue, which allowed the placement of the sample inside of the Omics pouch.
Table 1. Average murine tissue weights per condition, completed in triplicate.
| Tissue | Condition | Weight (mg) |
| Kidney* | Fresh | 235 |
| Kidney | Frozen | 289 |
| Heart | Fresh | 252 |
| Heart | Frozen | 188 |
*Two fresh kidney tissue repeats kept in MACS Tissue Storage Solution (Miltenyi Biotec) at 4°C for 24 hours.
Fig 2. Nuclei suspension preparation workflow using fresh or frozen tissue and the VIA Extractor™ tissue disaggregator in conjunction with the VIA Freeze™ Uno controlled-rate freezer.
Using a luer lock syringe, 4 mL of tissue lysis solution was added to each section of the Omics pouch and was secured by the Omics Clamp. Samples were processed at 200 rpm and 22°C for 15 minutes.
The contents of each Omics pouch portions were removed from the ports using a 5 mL luer lock syringe and subsequently filtered through a 50 μm cell strainer. Once centrifuged, the supernatant was removed. Then the pellet was resuspended in fixing solution and incubated on ice. A volume of ice-cold PBS was added to the suspension, mixed, passed through a 20 μm cell strainer, and centrifuged under the same conditions. Nuclei were resuspended in ice-cold PBS and counted on NucleoCounter NC-200 (ChemoMetec A/S) using Via2-Cassette (ChemoMetec A/S) and viewed using brightfield microscopy paired with a hemocytometer. A statistical analysis was completed using JMP 15.2 (SAS Institute).
Results
In the data shown below, we demonstrate that the VIA Extractor™ tissue disaggregator has adaptable capabilities not limited to live, single-cell dissociation but also single-nuclei isolation.
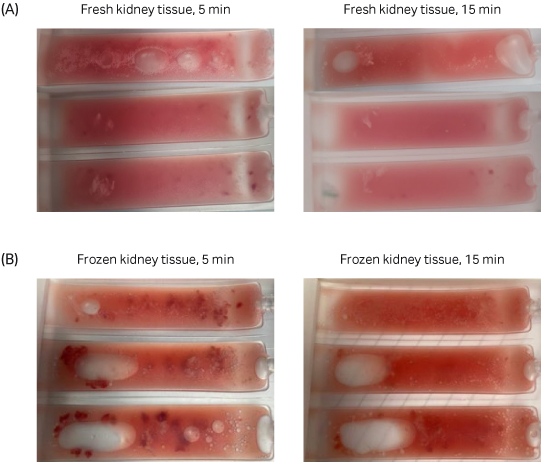
Fig 3. A comparison between fresh and frozen kidney tissue suspension in the Omics pouch 5 minutes into dissociation versus 15 minutes into dissociation. Both fresh and frozen kidney samples were completely dissociated after 15 minutes using the VIA Extractor™ tissue disaggregator.
Fresh and frozen samples were dissociated successfully after 15 minutes using the VIA Extractor™ tissue disaggregator at 200 rpm and 22°C (Fig 3).
Table 2. Average nuclei aggregation per tissue type and condition as a percentage with standard error values.
| Tissue | Condition |
Aggregation (% ± SE) |
| Kidney | Fresh | 2.3 ± 0.7 |
| Kidney | Frozen | 2.0 ± 1.0 |
| Heart | Fresh | 1.7 ± 0.9 |
| Heart | Frozen | 4.3 ± 2.4 |
To alleviate downstream issues while performing snRNA-seq, keeping aggregation to a minimum is key. Following dissociation of nuclei using the VIA Extractor™ tissue disaggregator, we observed minimal variability among the percentage aggregation between fresh and frozen kidney and heart samples, with all mean averages returning < 5% aggregates in each sample (Table 2; t-test; Kidney p = 0.80; Heart p = 0.39).
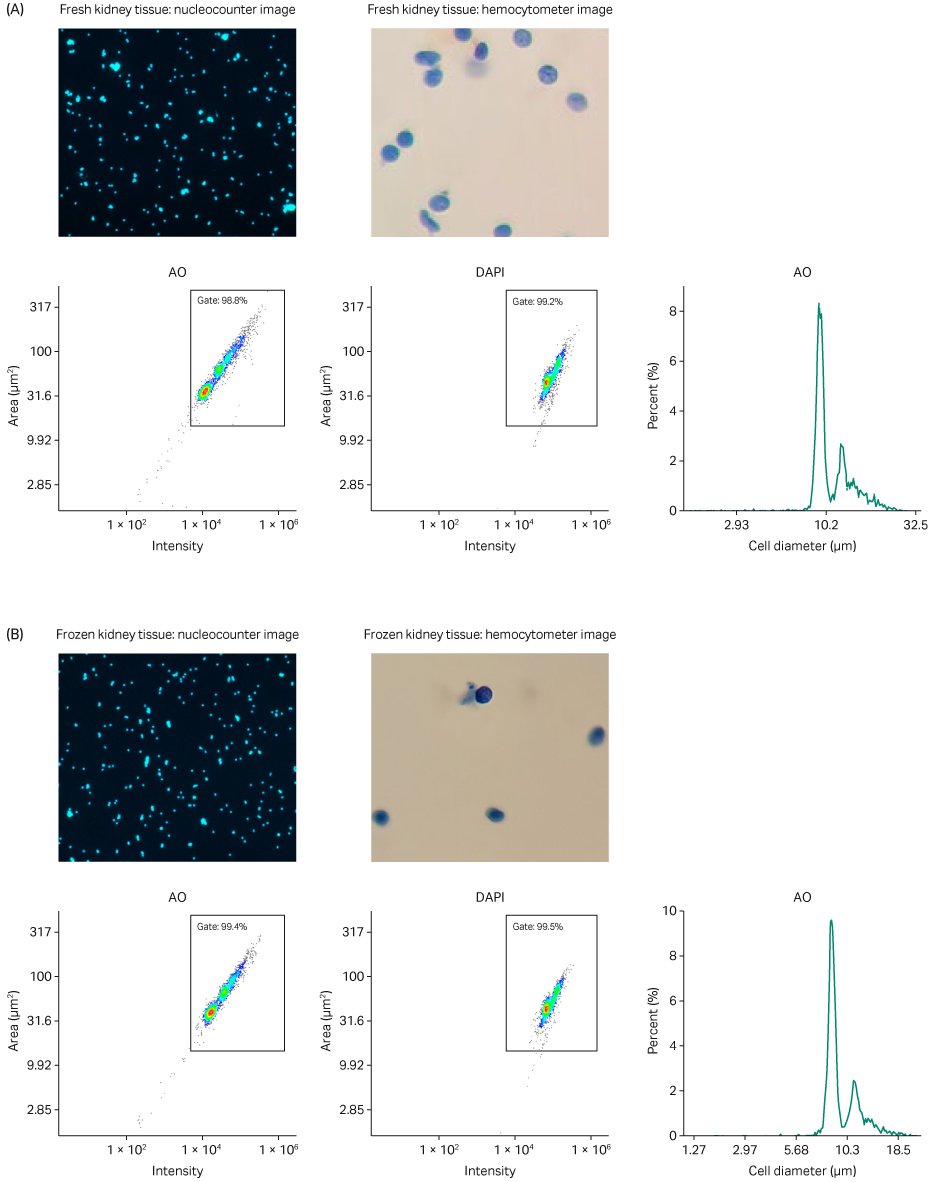
Fig 4. Analysis of single nuclei suspension from (A) fresh and (B) frozen kidney tissue. AO and DAPI merged staining using a NucleoCounter NC-200 and Via2-Cassette; nuclei-integrity check using brightfield microscopy paired with a hemocytometer; AO intensity versus area scatter plot with automatic gate; DAPI intensity versus area scatter plot with automatic gate; histogram representing estimated cell diameter versus percentage present in sample (cell diameter substituted for nuclei diameter as default).
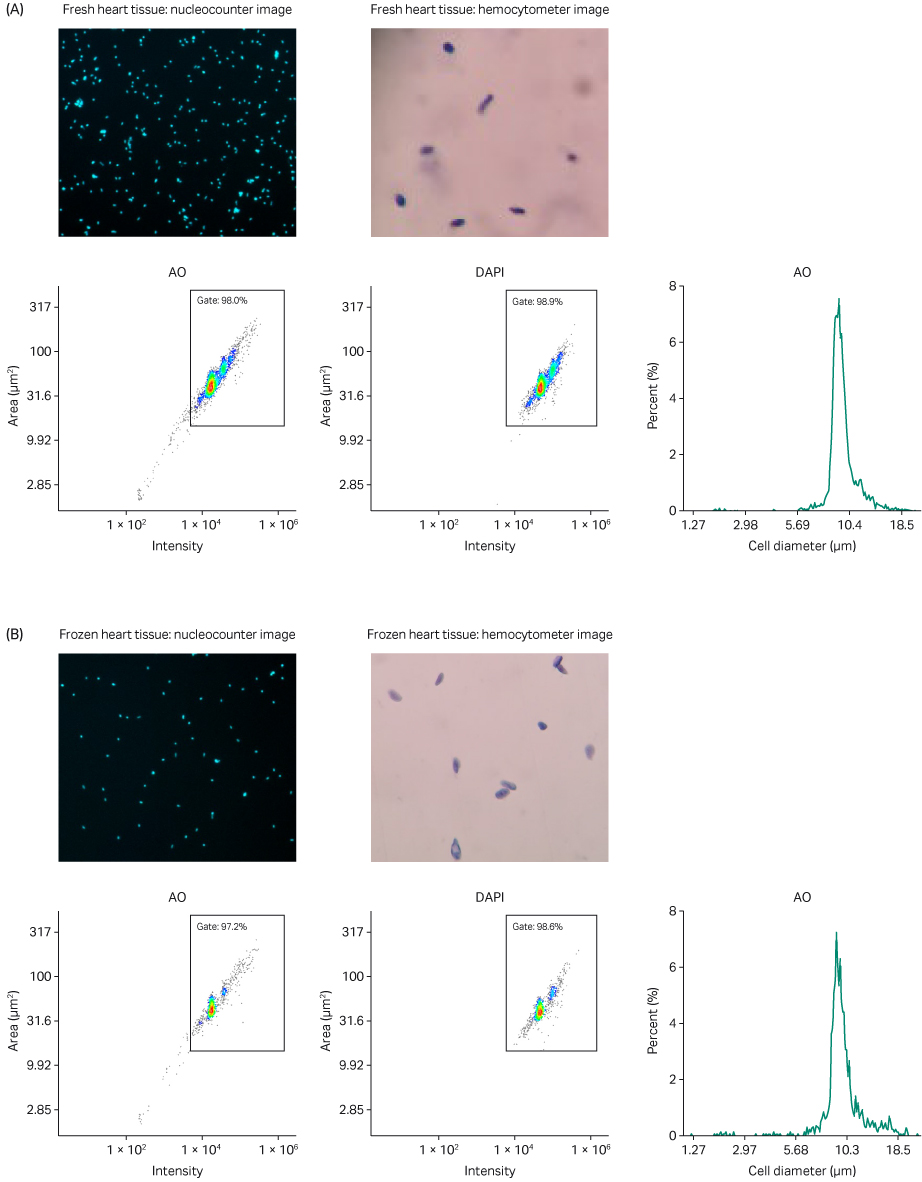
In conjunction with the automatically calculated numerical aggregation estimate, physical proof of minimal nuclei aggregates is shown in the acridine orange (AO) and 4’,6-diamidino-2-phenylindole (DAPI) merged staining (Figs 4 and 5). Because AO and DAPI both have a high affinity for DNA, the images show specific nuclei and their surrounding neighbors in sharp contrast. These data suggest that the VIA Extractor™ tissue disaggregator can reliably prepare nuclei suspensions with a low percentage of aggregates from both fresh and frozen kidney and heart tissue.
Using brightfield microscopy paired with a hemocytometer, we were able to assess the integrity of the nuclear membranes present in each sample. Each brightfield image suggests that the nuclear membranes remain intact (Figs 4 and 5). To further reinforce our observation, in the heart samples specifically, you can easily spot the characteristic elliptical shape of a cardiomyocyte’s nucleus (Fig 5). The VIA Extractor™ tissue disaggregator can process tissue and isolate single nuclei without causing damage to the nuclear membrane in kidney and heart tissues.
Fig 5. Analysis of single nuclei suspension from (A) fresh and (B) frozen heart tissue. AO and DAPI merged staining using a NucleoCounter NC-200 and Via2-Cassette; nuclei-integrity check using brightfield microscopy paired with a hemocytometer; AO intensity versus area scatter plot with automatic gate; DAPI intensity versus area scatter plot with automatic gate; histogram representing estimated cell diameter versus percentage present in sample (cell diameter substituted for nuclei diameter as default).
The scatter plots show the nuclei size distribution and staining intensity of AO and DAPI. Minimal small, dim particles are counted (bottom left of the scatter plots), which suggest that limited amounts of debris are present in both tissue types, regardless of whether the sample is fresh or frozen. The analogous nature of the heatmap-like plotted points within each gate across Figures 4 and 5 further confirm the presence of nuclei post-processing.
Each histogram (Figs 4 and 5) has a regular peak around a cell/nuclei diameter of approximately 9 to10 μm, which is consistent with the fact that a typical mammalian nucleus is approximately 5 to10 µm (4). The small peak to the right of the major peak in the histograms present in Figure 4 likely represents nuclear doublets, which are found at a much lower percentage of the total material present in the sample.
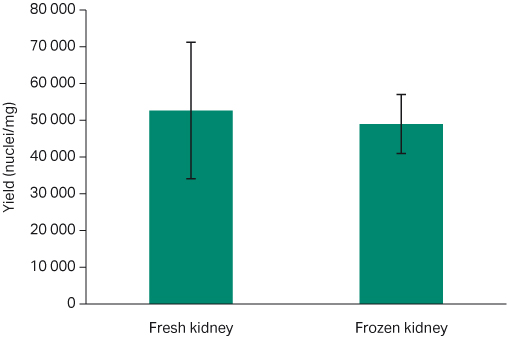
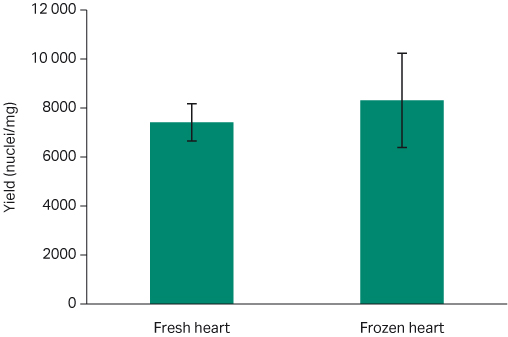
The typical nuclei recovery yield using the Chromium Single Cell Nuclei Isolation Kit (10X Genomics) from healthy frozen tissue ranges from approximately 5 to15 thousand nuclei/mg (5). Our results show proportional consistency between each tissue type, with no significant difference between fresh and frozen samples (P > 0.05). The fresh murine kidney samples yielded an average of 52 667 ± 18 559 nuclei/mg, while the yield for the frozen kidney tissue was 49 000 ± 8020 nuclei/mg (Fig 6; t-test p= 0.86). For fresh murine heart tissue samples, the average yield was 7414 ± 762 nuclei/mg; whereas for the frozen heart samples, the yield was 8316 ± 1926 nuclei/mg (Fig 7; t-test p=0.89).
Fig 6. Average nuclei yield per milligram comparison for both fresh and frozen kidney tissue with standard error bars. Fresh: 52 667 ± 18 559 nuclei/mg; frozen 49 000 ± 8020 nuclei/mg (t-test p= 0.86).
Fig 7. Average nuclei yield per milligram comparison for both fresh and frozen heart tissue with standard error bars. Fresh: 7414 ± 762 nuclei/mg; Frozen 8316 ± 1926 nuclei/mg (t-test p=0.89).
Conclusion
We previously demonstrated that the VIA Extractor™ tissue disaggregator can produce single-cell suspensions of high yield and viability with low cellular fragility.
Here, we show that the VIA Extractor™ tissue disaggregator can process solid kidney and heart tissue samples into single-nuclei suspensions. Whether the tissue sample is fresh or frozen, the VIA Extractor™ tissue disaggregator can produce consistent and homogenous single-nuclei suspensions with a high yield, intact nuclear membrane, and a low proportion of aggregates. The suspensions could be used further downstream for snRNA-seq and subsequent analyses to ascertain the transcriptional landscape of rare cell populations.
This data is based on three independent experiments with the equal number of replicates in each experiment. All samples tested were treated equally (with the number of replicates being the same for all products tested in the comparison) and according to manufacturers’ protocol and recommendations. Data was collected at Cytiva, Maynard Centre, Cardiff, UK (R&D Laboratory) during April to August 2022 and is held at this location.
References
- Wu H, Kirita Y, Donnelly EL, Humphreys BD. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: Rare cell types and novel cell states revealed in fibrosis. J. Am. Soc. Nephrol. 2018;30(1):23-32.
- Fish KN, Soderberg-Naucler C, Mills LK, Stenglein S, Nelson JA. Human cytomegalovirus persistently infects aortic endothelial cells. J. Virol. 1998;72(7):5661-5668.
- Condorelli G, Morisco C, Stassi G, et al. Increased cardiomyocyte apoptosis and changes in proapoptotic and antiapoptotic genes bax and bcl-2 during left ventricular adaptations to chronic pressure overload in the rat. Circulation. 1999;99(23):3071-3078.
- Li G-W, Xie XS. Central dogma at the single-molecule level in living cells. Nature. 2011;475(7356):308-315.
- What is the expected nuclei yield recovered using the Chromium Nuclei Isolation Kit. Accessed September 15, 2022.