High quality libraries are key to keeping next-generation sequencing costs down and maximizing usable data. Take a closer look at how size selection can improve your data quality, and the methods you can use in your library construction workflow.
NGS sample preparation, cost, and data quality
The introduction of next-generation sequencing (NGS) technology resulted in a fundamental shift in our approach to genomics. Even now, more than a decade after second generation sequencers arrived, the market continues to grow.
This is partly because of the constant drive to reduce the cost of sequencing and open up the technology to more researchers and applications. Despite these year-on-year cost reductions, individual sequencing runs remain expensive. Maximizing the usable data from any given run, which can be achieved by optimizing upstream library construction and sample preparation steps, can lead to additional savings.
These processes are relatively inexpensive and have substantial influence on final data quality. Here’s why library fragment size selection is a key step towards data quality, and my recommendations on the main methods for carrying out size selection, their advantages and disadvantages.
What does a typical NGS sample prep look like?
Although there are multiple approaches to sequencing, Illumina’s sequencing-by-synthesis approach continues to be the most widespread. We’ve previously discussed the fundamentals of NGS sample prep, which has several common steps for library construction, including:
- Fragmentation through enzymatic or mechanical means.
- End-repair and processing to homogenize the heterogeneous fragment ends.
- Adapter ligation for cluster generation and in-cell clonal amplification.
- Size selection to remove suboptimal fragment sizes and any adaptor dimers.
The significance of size selection
Genomic sequencing relies on having high quality libraries. Part of this is making sure library fragment sizes are within the optimum range for a given instrument, typically 200-500 bp for Illumina™ systems. This range is a consequence of the effect of fragment length on cluster generation and the efficiency of the sequencing process itself.
Small fragments tend to cluster more efficiently on the flow cell than larger fragments. A bias towards smaller fragments leaves much of the sequencing capacity unused. Selecting fragment sizes below 150 bp can risk carryover of unwanted adaptor and primer dimers, the sequencing of which leads to a lot of unusable data and further wasting of capacity.
Fragments larger than optimum pose the opposite challenge. Although it’s possible to sequence fragments >1 kb in length, this is inefficient and prone to errors—an issue that third generation sequences attempt to solve.
Individual samples might also have different shearing profiles, with narrow to wide distributions. Setting an instrument up for 600 bp fragments when there is a 200-1,000 bp distribution, for example, means that many of the sequencing templates won’t be viable or read to sufficient depth. This produces little useful data and low uniformity of coverage.
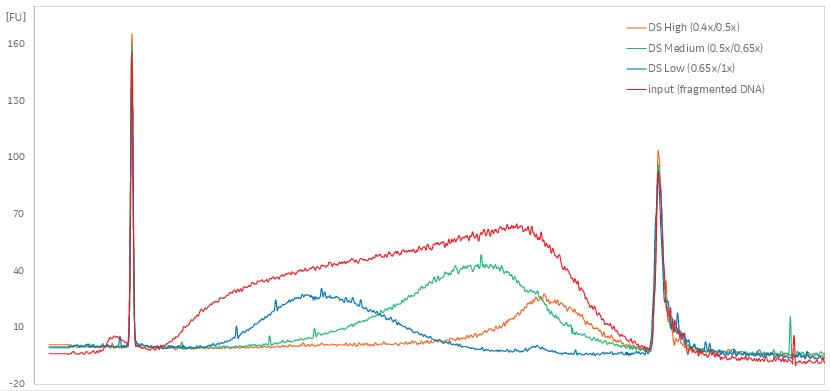
A size selection step enables you to take a randomly fragmented library and pull out only the fragments fitting the optimal/target range for the instrument and application (Fig 1). This saves time and cost by maximizing the efficiency of sequencing runs.
A note on DNA fragmentation methods
There are various options for fragmentation, some of which attempt to bypass the need for size selection altogether. The choice of method may depend on your application, starting material, and equipment available.
Enzymatic methods tend not to be completely random, but provide some control over fragment sizes through varied incubation times. However, these are less well suited for de novo assembly due to the likelihood of making fewer overlapping fragments.
There are various options for mechanical shearing, which use sonication or focused acoustic technologies. These are random, and can be tuned to produce predictable shearing profiles.
Size selection methods
The approaches to size selection include enzymatic, gel-based, and magnetic bead-based methods, the suitability of each depending on the needs of the experiment. These also provide an opportunity to clean up adaptor dimers and any other leftover reagents.
Enzymatic approach
Illumina’s Nextera™ kits produce libraries for various applications compatible with Illumina technology in one step.
When launched, they attempted to get around the need for size selection by using transposon-based fragmentation and tagging, known as ‘tagmentation’, saving several workflow steps. However, library profiles tended to be broad, leaving users often reverting to a separate size selection step.
Nextera kits now include magnetic bead-based size selection reagents.
Gel-based approach
Gels have long been used for nucleic acid purification, enabling you to physically remove the chosen fragment size. Gel-based systems, such as Sage’s Pippin Prep™, help automate this process, but have inherently limited throughput. A typical 96-sample batch requires close to 10 hours to process.
Magnetic bead-based approach
The introduction of magnetic beads for convenient and high throughput size selection and clean-up has transformed NGS workflows, with Cytiva’s Sera-Mag particles integral to this success.
Originally developed for the isolation of PCR products, these beads have polystyrene cores covered in magnetite and a layer of carboxyl molecules. Nucleic acids bind to them reversibly in the presence of polyethene glycol (PEG) and salt; a process known as solid phase reversible immobilization.
The beads are otherwise inert and have high binding capacities, due to large surface areas. The size of fragment bound can be adjusted by simply altering the volumetric ratio of PEG/salt/beads to DNA. From a practical point of view, this bead chemistry makes it straightforward to size select a very specific range of fragments consistently and reproducibly.
The magnetic bead-based approach is well suited for high throughput applications with automation, and the cost of reagents is also low compared to other approaches. These properties make magnetic beads a simple solution for optimizing NGS sample prep.
At Cytiva, our genomics experts can support you in optimizing your NGS workflow. To find out more about using magnetic bead-based size selection, or for help and advice on any aspect of your workflow, contact Cytiva Scientific Support. Or read our other blogs on NGS sample preparation.