Explore the challenges of DNA extraction (and RNA extraction) from a variety of common sample types, and the methods and technologies that facilitate this, including magnetic beads, which provide a versatile, high capacity solution.
Analyzing nucleic acids is enormously powerful, providing us with insight into a variety of biological processes for basic research and clinical applications. DNA isolation (and RNA isolation) is the first step for many modern genomics techniques and applications, which require high-quality starting material free of contaminants.
For lab managers complexity remains at the heart of nucleic acid extraction. You could say there are both too many and too few choices out there. What is the ‘right’ isolation protocol for your sample or application?
In this blog, I’ll explore the challenges of isolating nucleic acids from a variety of common sample types and pick out several approaches, highlighting magnetic bead DNA extraction, which has become one of the most versatile methods for nucleic acid isolation.
How to extract DNA (or RNA)
Genomic DNA extraction is the first step in many molecular biology studies, and all recombinant DNA techniques. Protocols involve breaking open the cells and separating the DNA you need from other nucleic acids and cellular components in the sample, while also keeping it in good condition for downstream analysis.
There are several approaches that you might take, varying from gentle to aggressive. The choice depends on several factors, including the target DNA, source organism, the type and quality of your starting material, and the application. They generally all share three common steps: lysis, contaminant removal and DNA recovery.
Step 1: Lysis
Cell lysis involves chemical, mechanical, or enzymatic disruption of cell membranes and denaturation of proteins. The exact method depends on your starting material. Bacteria, mammalian cells, plant cells, and human tissues all might require a slightly different approach.
‘Gentle’ lysis might involve using a detergent, such as sodium dodecyl sulfate (SDS), or enzymes to break up cell membranes; aggressive lysis might take the form of homogenization to physically break open cell walls.
Step 2: Removing contaminants
You can use both solution-based and solid-phase methods to separate DNA from unwanted lysis debris and potential contaminants. Phenol chloroform DNA extraction, for example, separates water-soluble DNA and denatured proteins into different phases. This is cheap, but slow, and risks carryover of phenol that can affect downstream applications.
Solid-phase extraction binds DNA to a column or bead surface. Silica resins or silica-coated magnetic beads, for example, use chaotropic salts to disrupt hydrogen bonds and bind nucleic acids, enabling contaminants to be washed away. Oligonucleotide-coated resins can also add a level of specificity, but column kits can quickly add up in cost.
Step 3: Recovering the target nucleic acid
Downstream applications require your DNA in a suitable format (solvent and concentration). Often, this will be just a matter of precipitating your DNA with ethanol, washing, and resuspending in an appropriate buffer. For solid-phase methods, it will first require adjusting the pH or salt concentration of the buffer to release the nucleic acids.
Sample-specific DNA isolation challenges
Cultured mammalian cells and tissues
Cultured cells are relatively easy to lyse with osmotic shock or detergent treatments, while isolating DNA from tissue requires breaking down the extracellular matrix, not just cell membranes. This often requires homogenization followed by silica column (e.g. illustra kits) or mag bead-based (e.g. SeraSil-Mag) purification, or less favorable phenol-chloroform extraction.
Using formalin-fixed, paraffin-embedded (FFPE) tissue is common in clinical applications and some research studies. It’s excellent for preserving tissue structures, but can introduce all sorts of DNA damage with profound effects. That is, as the quality of the DNA isolated directly affects the assay results, positive samples might be overlooked simply because of poor extraction.
Blood
A challenge of DNA extraction from blood is the variability in DNA quantity depending on blood fraction. Red blood cells don’t contain DNA, so there’s much less per cell in whole blood compared to buffy coat or bone marrow-derived fractions.
Blood coagulation also presents challenges: clotting can prevent effective sample digestion, and some anticoagulants can interfere with PCR amplification.
Bacteria
There are differences between gram-positive and gram-negative samples in DNA extraction from bacteria. Gram-positive samples usually require lysozyme treatment to digest the higher levels of peptidoglycan in the cell wall, whereas for gram-negative samples, a simple osmotic shock might be enough.
DNA is unlikely to be scarce with either type, and it’s common to use fast methods, like alkaline extraction and diatomaceous earth, to extract the DNA. Both methods are reliable, but alkaline extraction might not provide the highest purity by itself, and diatomaceous earth can be high cost.
Plant material
Plant cells can be embedded in a tough matrix and have cell walls consisting of glycans and cellulose that are difficult to break. The solvent-based cetyltrimethylammonium bromide (CTAB) extraction method is common for plant material, but it is an aggressive approach. It uses harsh chemicals, is laborious, and often requires further clean-up and optimization for different samples and applications.
To bead or not to bead for DNA extraction
Magnetic beads provide an excellent alternative to traditional isolation and clean-up methods due to their versatility and ease of use. They don’t require additional centrifugation of a potentially already agitated sample, improving the likelihood of recovering larger fragments, and can be scaled up to have a higher binding capacity than columns.
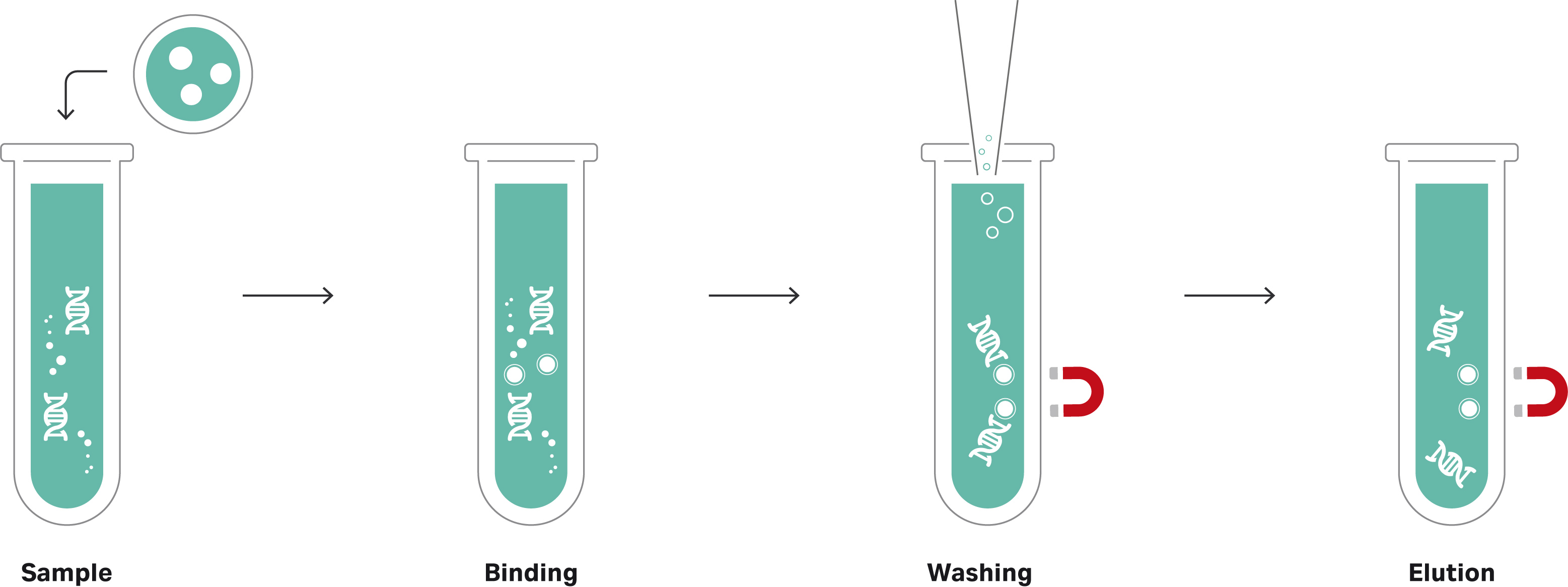
Using magnetic beads is straightforward, needing no hazardous solvents, and releasing the DNA or RNA is just a matter of adjusting the buffer properties (Fig 1). This simplicity also makes magnetic beads well suited to automation in high-throughput applications.
Fig 1. The principle of magnetic beads for nucleic acid isolation.
Cytiva's SeraSil-Mag silica coated magnetic beads are an appropriate example. They help address several challenges in DNA extraction and clean-up I’ve mentioned here, and suit a range of applications, including all the sample types I’ve described, when used with appropriate buffers. Their binding capacity and tight size distribution deliver highly reliable results while being easy to use without centrifugation.
At Cytiva, our genomics experts aim to support you in all your workflows. Review our magnetic bead product range or check out our other genomics blogs to find out more about optimizing your workflows. For support in any aspect of your application, contact our Scientific Support team or your local Cytiva representative.