Explore the mRNA universe with Cytiva. In this series, our scientists share their journey and insights on mRNA synthesis for research purposes. And they share their advice on how to scale. Through peer-to-peer interactions, we continue to encourage the spirit of collaboration developed at a time when new modalities are being used as therapies for the first time.
Part 2 – DNA template
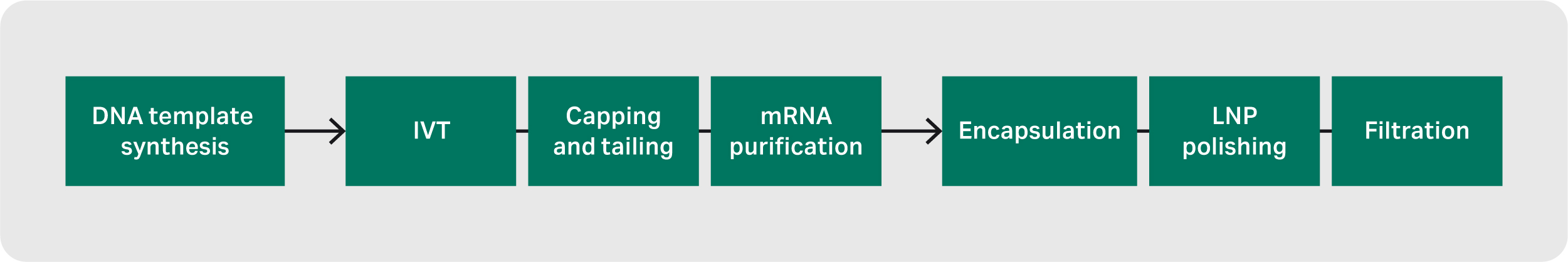
The creation of an encapsulated mRNA–LNP begins with synthesizing the DNA template. If plasmid DNA (pDNA) is used as the template, additional steps are necessary, such as purification after host cell lysis and linearization of the circular pDNA. The DNA template then undergoes purification prior to in vitro transcription (IVT), during which mRNA is formed.
Capping of the 5’ end of the mRNA takes place either during or after IVT and the addition of the polyA tail is usually included in the DNA template. Impurities from the IVT reaction are removed, and the purified mRNA can be sterile-filtered before being encapsulated into a lipid nanoparticle (LNP). Finally, the mRNA–LNP is polished and sterile-filtered (Fig 1).
Fig 1. The process of creating an encapsulated mRNA–LNP.
DNA template synthesis
DNA template containing the sequence to be transcribed with an upstream RNA polymerase promoter site serves as starting material for IVT. The template can be derived in several ways, such as from pDNA or by a polymerase chain reaction (PCR) product. The choice depends on the conditions of the plasmid and intended application, and there are benefits to each option. The pDNA route generates many copies of the same template by hijacking the bacteria’s machinery. If the DNA template is derived from plasmids, the pDNA should contain your gene of interest, RNA polymerase promoter site and untranslated regions (UTRs), a unique restriction enzyme cut site for linearization downstream of the 3’ UTR or polyA tail, origin of replication, polyA tail, and a gene for antibiotic resistance for use as a selectable marker. If elements such as a polyA tail and/or RNA polymerase promoter sequence are missing in the plasmid, a PCR product may be preferred since it is possible to design such sequences in the primers.
The endotoxin-free PCR method offers greater flexibility, creating different templates in a shorter amount of time since there are no incubation and linearization requirements. On the other hand, it is a highly viable solution if only small amounts are needed, since it is more expensive to perform at scale.

Generating a DNA template using PCR
PCR amplifies the DNA sequence of interest by using a reaction mixture consisting of the DNA sequence, primers, DNA polymerase, deoxyribonucleotides (dNTPs), and PCR buffer, and then using thermal cycling to help the replication process. After the reaction, the PCR product is readily purified using a PCR purification kit. PCR allows the conversion of any DNA fragment to a transcription template by affixing an RNA polymerase promoter (e.g., T7) to the forward primer.
PolyA-tailed reverse primers can be used during PCR to generate transcription templates with polyA tails, obviating the need for an additional polyadenylation step after transcription. These reverse primers are particularly useful for screening different polyA tail lengths, especially for short mRNAs. For long mRNAs, post-transcriptional polyadenylation is a better option since the reverse primers can be very long and may be more costly. Although phenol-chloroform extraction or ethanol precipitation are purification methods traditionally used in the laboratory, incomplete removal of organic solvents will cripple subsequent enzymatic steps. For example, these solvents can denature enzymes, leading to failed IVT reactions or low mRNA yield. Scalability is also an issue.
To determine DNA quantity and purity, UV absorption (A260/A280) using a spectrophotometer is commonly used. Agarose gel electrophoresis, sequencing using primers, or bioanalyzers can be used to check the integrity of the DNA template.
Challenges
- DNA template construction is a labor-intensive and time-consuming process.
- Linearization (pDNA only) and purification of the DNA template can result in variable purity and quality.
- DNA template quality affects transcription yield and the mRNA integrity. The higher the purity, the greater the transcription yield.
- Handling of organic solvents (phenol and chloroform) and their presence (if not fully removed) denature linearization restriction enzymes and IVT enzymes.
- It is difficult to maintain a nuclease-free environment and acquire high quality nuclease-free reagents (enzymes, nucleotides, and primers) for PCR and sequencing.
- There is a need for simplified, lab-friendly preparative chromatography steps and resins for larger-scale purification.
Strategies
- Aldevron offers high quality standard and linearized pDNA, ranging from microgram to multi-gram scales, complete with Certificate of Analysis.
- illustra™ plasmidPrep kits from Cytiva simplify pDNA extraction and purification.
- The Mini Spin Kit can yield 6–15 µg of purified pDNA from 1.5–3 mL E. coli cultures for molecular cloning (A260/A280 > 1.8) in less than 10 mins without the use of organic solvents.
- For larger culture volumes and transfection quality (A260/A280 = 1.8–2.0) pDNA, the Midi Flow Kit can yield 20–250 μg from 25–150 mL culture medium.
- The Cytiva illustra™ GFX PCR DNA and Gel Band Purification Kit is ideal for DNA volumes from PCR (up to 100 μL) or from gel slices (up to 900 mg), purifying DNA ranging from 50 bp up to 10 kbp in size.
- If handling large numbers of samples in solution and DNA 100 bp–10 kbp in length, the illustra™ GFX 96 PCR Purification Kit allows purification of up to 96 PCR products simultaneously in 15 min.
- Cytiva and Pall Corporation offer solutions for purification for gene therapy and vaccine applications.
- Anion exchange chromatography: Mustang® Q membrane absorbers from Pall Corporation are used in this pDNA capture step with process intensification and overall recovery in mind. These membrane absorbers are multi-layered, compact, and possess high dynamic capacity. Within the Mustang® Q family, the Mustang® Q XT Acrodisc Units are fit for laboratory scale. The membrane volume of < 1 mL reduces the amount of sample required for evaluation, and the female Luer lock inlet and outlet simplifies connection to typical low-pressure chromatography systems.
- Hydrophobic interaction chromatography: Capto™ PlasmidSelect resins from Cytiva are involved in this isoform separation step. These resins allow high-flow separation of supercoiled, covalently-closed, circular forms of pDNA from open circular forms and are available prepacked in HiTrap™ and HiScreen™ columns from Cytiva. In addition, these resins can be used to determine pDNA concentration and analyze the ratio of supercoiled to open circular pDNA.
- Cytiva offers high quality nuclease-free PCR and sequencing reagents.
- In addition to RNaseAlert® kit and Nuclease Decontamination Solution, IDT also provides nuclease-free water and custom primers.
In part 3, we will look at challenges and strategies related to the mRNA product IVT process.
“In vitro transcription, or IVT, is an enzymatic reaction. In our lab, we’ve found that IVT is very sensitive to the quality and concentrations of the ingoing components, such as the plasmid template.” — Susanna Lindman, Principal Scientist at Cytiva R&D, Uppsala, Sweden