Enzymes play key roles in many applications, including NGS. See how a range of enzymes from your molecular biology toolbox make the NGS library preparation workflow easier.
Back to basics: what are enzymes and what do they do?
Enzymes are macromolecular biological catalysts (usually proteins) that accelerate virtually all chemical reactions in cells. They work by reducing the activation energy needed for reactions that might otherwise take a long time, or not take place at all.
An enzyme’s active site binds one or more substrates in such a way that it minimizes the necessary energy input. Molecules enter the active site, and the enzyme facilitates the chemical reaction. The speed of these reactions is determined by the action of the enzyme.
Some enzymes bind a range of substrates while others are quite selective, either through the shape of the active site or associated targeting molecules, like oligonucleotides.
In next-generation sequencing (NGS) sample and library preparation , you probably use enzymes at almost every critical step. They can digest tissue and other unwanted cellular material, degrade or reverse transcribe RNA, amplify starting material, cut DNA to optimum fragment lengths. The list goes on.
The enzymes in NGS library prep
A typical NGS library preparation workflow has multiple steps, several of which involve enzymes:
- Extracting and purifying DNA (or RNA) from a sample
- Fragmenting DNA (if necessary)
- Size selecting fragments of optimum lengths for sequencing
- Fragment end-repair
- Ligating adaptors
- Amplifying the library (if necessary)
- Quantitating and pooling for sequencing
Using enzymes for DNA extraction
Getting a purified sample can be a messy business. Depending on the starting material, you might have challenges with yield, integrity, and purity.
Tissue samples, for example, might be a plentiful source of nucleic acids, but preservation methods can damage DNA. They also often need homogenization to break down the extracellular matrix for DNA or RNA extraction, risking further unwanted damage and fragmentation.
Single-cells, on the other hand, provide a limited yield. The nucleic acids require amplification to be usable for NGS or any other genomics application.
These are both situations where enzymes can be useful. Proteases can help degrade nucleases and other proteins, though it’s also common to have DNases or RNases in lysis buffers to degrade the unwanted nucleic acid type.
Commercial kits containing cocktails of repair enzymes can restore damaged DNA, such as that from formalin-fixed paraffin-embedded (FFPE) samples, to a more useful state.
When you’re dealing with limited or insufficient yields, where extraction alone is impractical, whole genome amplification (WGA) can help enable otherwise impossible analyses. Using Phi29 DNA polymerase and random primers for multiple displacement amplification (MDA) generates micrograms of high molecular weight DNA from picograms of starting material, providing a simpler alternative to PCR-based methods.
Library fragmentation
Part of generating a high-quality library for NGS involves making sure fragment sizes are within a range centered around an optimal average length, typically a few hundred base pairs for Illumina™ systems.
There are several options for DNA fragmentation, including mechanical, enzymatic, and chemical approaches. Mechanical methods might involve hydrodynamic shearing by sonication, focused acoustic shearing, or nebulization. But each comes with caveats:
- Sonication is easy and effective, but requires careful calibration and can be slow compared to other methods.
- Focused acoustic shearing provides tight size distribution, but requires specialized equipment with potentially high upfront costs.
- Nebulization is fast, but produces a rather wide size distribution and degrades much of the sample, and so requires high input.
All mechanical methods also have the potential to introduce unwanted DNA damage as they won’t necessarily make clean breaks.
Enzymes provide a scalable and cost-efficient alternative. They don’t require any complex equipment, meaning upfront costs are low. Enzyme reactions are generally quite gentle reactions compared to mechanical shearing, minimizing unwanted sample degradation or DNA damage.
Reaction volumes can also be as small as a few microliters, and there’s little risk of losing a precious or limited sample.
The enzymes used for DNA fragmentation fall into three types: restriction enzymes, nicking enzymes, and transposases:
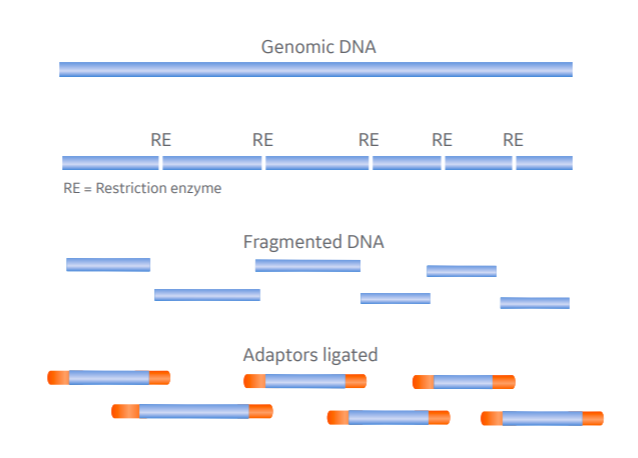
1. DNA fragmentation: restriction enzymes
Restriction enzymes (or restriction endonucleases) are essential tools for all labs working with recombinant DNA. There is a large variety available, typically targeting 4 to 8 base pair sequences, and they serve a useful function in NGS library prep.
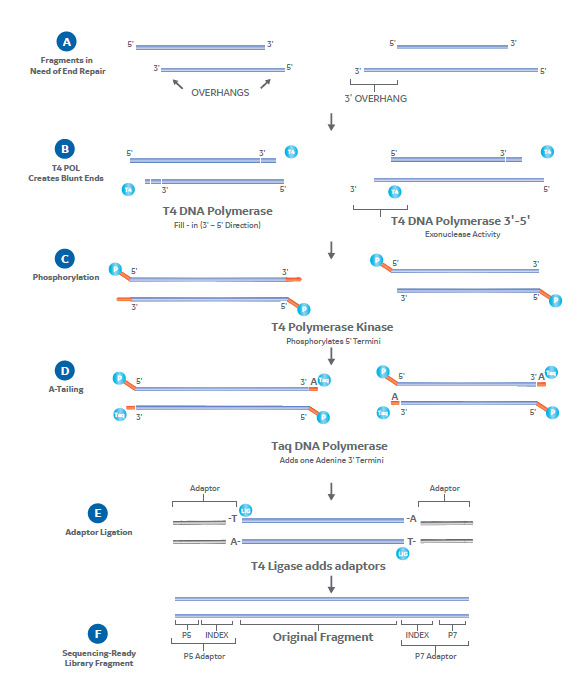
On binding their recognition site, restriction endonucleases create either blunt-ended or overhanging double-stranded breaks, the latter requiring end-repair by fill-in (e.g. by the Klenow fragment of DNA Polymerase I) before adaptor ligation (Fig 1A).
By selecting an enzyme with a recognition site that appears roughly as often as the target average fragment length, you stand the best chance of creating an even distribution of fragments for NGS.
But these recognition sites are also the main drawback of restriction digests. They will introduce some fragmentation bias (i.e. fragmentation is not random). This isn’t an issue in all DNA sequencing applications but means some genomic regions might have lower coverage than others, a challenge for applications reliant on deep sequencing.
A
B
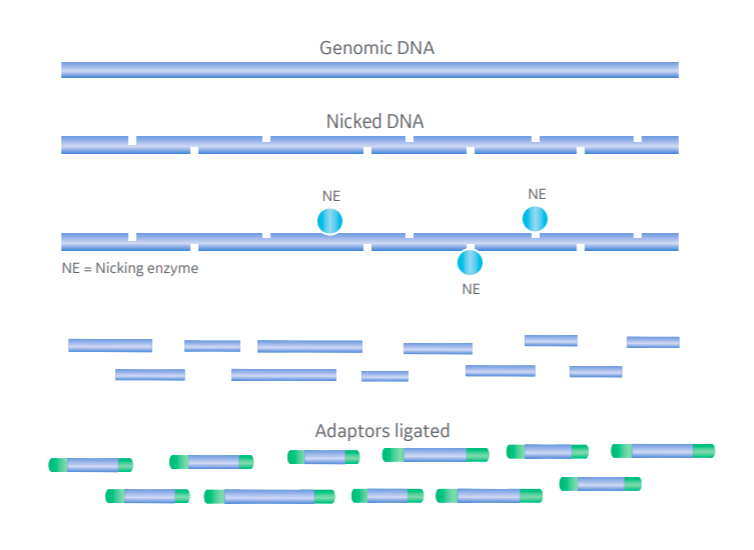
Fig 1. Comparing restriction endonuclease- and nicking enzyme-based DNA fragmentation in NGS. Genomic DNA fragmented at specific recognition sites by restriction enzyme in one step (A) and random sites by nicking enzyme and single-strand-specific endonuclease in two-step process (B).
2. DNA fragmentation: nicking enzymes
An alternative to the potentially biased fragmentation introduced by restriction enzymes is the use of nicking enzymes. DNase I, for example, can make random single-stranded cuts in the DNA. A second, single-strand-specific enzyme that recognizes nicked sites then cleaves the second strand.
The result is a distribution of fragments with short overhangs that need fill-in before adaptor ligation, but are otherwise unbiased (Fig 1B). There’s also potential here for modulating the average fragment length by varying reaction conditions and time.
3. DNA fragmentation: transposomes
Whereas using restriction and nicking enzymes depends on cutting specific or random sites in the genome and performing end-repair, transposon-based fragmentation can both cleave DNA at random sites and insert a short double-stranded oligonucleotide on both ends. These are then ready for index and adaptor ligation without further processing.
Illumina’s Nextera™ kits use this ‘tagmentation’ approach to produce libraries compatible with Illumina technology in one step.
But one challenge with the use of transposases is that the reaction (and so the quality of the library) is sensitive to the amount of starting material. Each transposase only works once, so the average fragment length is critically dependent on the DNA:transposome ratio, though this does provide a way of modulating the target fragment length too.
Enzymes in DNA fragment end-repair and adaptor ligation
Adding adaptors to library fragments first requires clean blunt ends with a single-nucleotide 3’ A-tail amenable to ligation. As mechanical shearing and enzymatic methods tend to create damaged ends or overhangs, most tend to need repair before ligating adaptors. This doesn’t apply to Illumina’s ‘tagmentation’ method though, which fragments and adds short blunt-ended oligonucleotides as part of the same step.
Enzymes are key to end-repair. A typical blunting enzyme mix might, for example, contain T4 DNA polymerase and T4 polynucleotide kinase (PNK). T4 DNA polymerase (in the presence of dNTPs) can fill-in 5’ overhangs and trim 3’ overhangs down to the dsDNA interface to generate the blunt ends (Fig 2A-B). The T4 PNK can then phosphorylate the 5’ terminal nucleotide (Fig 2C).
A-tailing also requires a polymerase. Taq DNA polymerase the most common as it has terminal transferase activity and naturally leaves a 3’ terminal adenine (Fig 2D). DNA polymerase I Llarge (Klenow) fragment is another common option, which can also double as a blunting enzyme. Using either of these polymerases leaves A-tailed ends that complement standard Illumina sequencing adaptors.
Adding an adaptor at this stage just requires an incubation with T4 DNA ligase . This enzyme will join both blunt and so-called ‘sticky’ ends, in this case catalyzing the formation of a phosphodiester bond between the 5’ and 3’ termini of the end-repaired fragments and sequencing adaptors (Fig 2E-F).
Fig 2. End-repair and adaptor ligation in NGS library preparation. Fragments with 5’ and 3’ overhangs (A) are filled-in by T4 DNA polymerase to create blunt ends (B). T4 PNK then phosphorylates 5’ termini (C) and Taq DNA polymerase A-tails 3’ termini (D), leaving ends amenable to adaptor ligation. T4 ligase adds sequencing adaptors (E) to leave complete sequencing-ready library fragments (F).
Each of these steps must be accurate and efficient for the library to produce reliable NGS data, and so relies on using high-quality enzymes under optimum reaction conditions. The result is a library of fragments that might need quantitation and pooling, but is otherwise ready for sequencing.
Enzymes in DNA amplification
The last step in NGS library prep that might involve enzymes is amplification by PCR. This step isn’t always necessary. High-quality samples producing high yields or single-cell samples already amplified by MDA are unlikely to need a further amplification step.
PCR does serve a dual purpose though. As well as amplifying, in some protocols it’s needed for adding the functional elements. For example, Illumina’s ‘tagmentation’ approach has a reduced-cycle PCR step for adding sequencing adaptors. The adaptors bind and extend from the short oligonucleotides at the ends of fragments.
Whether you’re using PCR for adding indexes, or also for amplification, it’s essential that it be error-free. That requires using a particularly high-fidelity thermostable DNA polymerase with excellent proof-reading capabilities and appropriate reaction conditions. This approach maximizes amplification efficiency, and minimizes both amplification bias and the risk of introducing sequencing artifacts.
These factors combined ultimately help generate uniform coverage, even across high G-C and other difficult regions.
So, as you can see, enzymes are crucial to multiple steps in NGS sample and library preparation. They provide shortcuts for slow or difficult reactions, and enable us to modify, repair, and amplify nucleic acids for a variety of applications, particularly NGS.
At Cytiva, our genomics experts can support you in optimizing your NGS workflow. Find out more in our other NGS blogs , browse our products for molecular biology applications , or for help and advice on any aspect of your workflow contact Cytiva Scientific Support.