When you put your affinity-purified protein in the freezer, it seemed to be in solution. But after a few freeze-thaw cycles it looks a bit cloudy. Has your treasured protein aggregated? Maybe not. You might be able to recover a good portion of the remaining functional protein. Find out how.
Preparative size exclusion chromatography (SEC) is a powerful method for obtaining size-homogeneous protein. The following example describes a simple method for preparing milligrams of monomeric, functional protein. This method uses one of our highest resolution SEC columns, Superdex 75 Increase. The starting material was a preparation of partially purified histidine-tagged (his-tagged) protein that had oligomerized during storage and freeze-thawing.
First, purify your protein preparation with preparative SEC
Before starting, ensure that the selected SEC column is capable of providing the resolution required to separate your functional protein from oligomers. If you need to concentrate your protein sample, we recommend using a Vivaspin protein concentrator. If the protein has visibly precipitated, the sample should be cleared through a filter that has low protein binding. We recommend Protein Prep filters for ÄKTA Chromatography systems (0.45 µm).
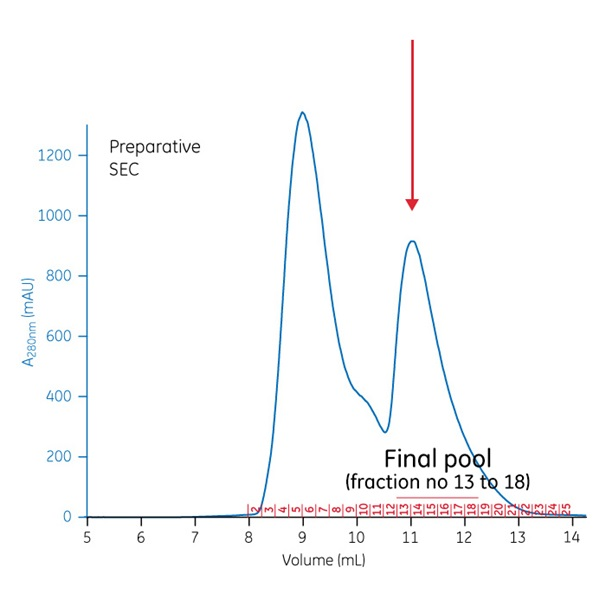
In our example we used an ÄKTA pure 25 chromatography system for purification. A 500 µL sample of the oligomerized protein preparation was loaded onto a Superdex 75 Increase 10/300 GL column. The flow rate was low (0.2 mL/min), and small fraction sizes (0.25 mL) were collected.
The preparative run gave essentially two peaks; the desired peak is the one that eluted later. It is marked on this chromatogram with an arrow. The fraction numbers are also shown in red.
Next, check samples of the fractions with analytical SEC
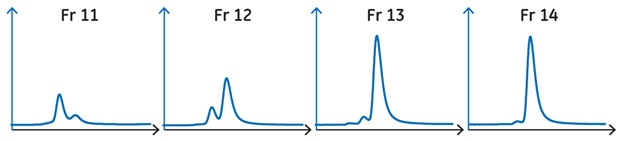
A benefit of SEC analysis is that it can reveal size-heterogeneity not visible with SDS-PAGE, because the SDS generally dissociates any noncovalent dimers and oligomers. Before deciding which fractions to pool, analyze samples of the fractions from the desired peak that are closest to the contaminant peak(s).
In our example, we used an ÄKTA pure 25 system along with a shorter, thinner version of the column used for preparative purification. The short column enabled us to analyze each sample in 6 min. Samples (25 μL) of the freshly collected fractions at the start of the desired peak were loaded individually onto a Superdex 75 Increase 5/150 GL column and run at 0.45 mL/min.
Based on SEC analysis fraction 13, but not fraction 12, was sufficiently size-homogeneous to be included in the pool.
Finally, pool the selected fractions and analyze with SEC
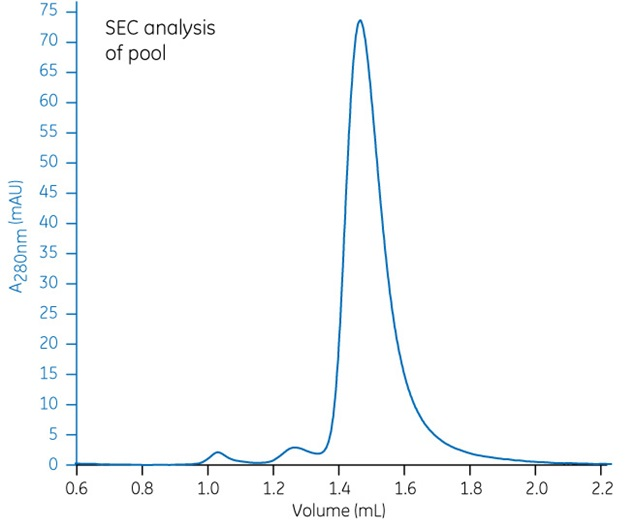
Fraction numbers 13 to 18 were pooled, and a 25 µL sample was analyzed with the same Superdex 75 Increase 5/150 GL column used in the previous step. The run was performed on an ÄKTA pure 25 chromatography system at a flow rate of 0.45 mL/min.
Approximately 5 mg of highly size-homogeneous target protein was obtained using this method. The difference between the sample before and after SEC purification is significant. Download the Superdex 75 Increase Data file and check out the SDS-PAGE analysis results for yourself.
Superdex 75 Increase columns are designed for separation of proteins and other biomolecules from Mr 3000 to 70 000, which makes them suitable for many recombinant tagged proteins. Three column sizes are available for small-scale preparative and analytical SEC.