How and why do you normalize your NGS libraries? Read up on why we need to normalize, best practices for quantitating your libraries (hint: use qPCR), and how magnetic beads enable a new approach to normalization.
What is DNA normalization and why is it important in NGS?
Normalization in next-generation sequencing (NGS) is the process of equalizing the concentration of DNA libraries for multiplexing. Multiplexing helps maximize the use of the ever-increasing capacity of NGS technology, enabling you to run multiple—often thousands—of libraries on a single flow cell, and drive down costs.
These ever-reducing DNA sequencing costs have led to its adoption in an array of molecular diagnostics applications, including reproductive health and oncology, as well as enabling a range of clinical research initiatives around the world.
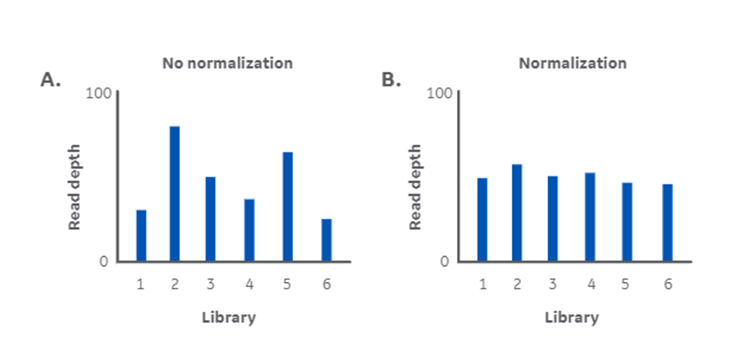
Both basic research and clinical NGS rely on obtaining reliable data. But uneven library concentrations from different types and qualities of sample can lead to inconsistencies in data quality (Fig 1).
Those libraries with a high concentration are likely to be overrepresented on the flow cell while those with low concentration are underrepresented. Overrepresentation isn’t necessarily a problem, likely increasing read depth, though it does waste capacity. Underrepresentation might result in poor read depth and unreliable data, wasting capacity and potentially your precious sample.
This highlights the importance of normalization in making sure every library is represented equally and sequenced to sufficient depth (Fig 1).
What are the implications of normalizing and not normalizing libraries?
From a cost standpoint, wasting capacity means you end up spending additional work time re-preparing libraries, assuming there is sample available. This time could be better spent on downstream analysis or preparing the next batch of libraries.
From an application and outcome standpoint, analyses and decisions based on potentially inaccurate or incomplete data will at best confuse research results or lead to repeating experiments. At worst, clinicians might, for example, miss key information like a rare allele or single nucleotide variation (SNV) that could have led them down a more appropriate treatment avenue.
Normalization helps address these challenges.
Fig 1. DNA library normalization addresses the challenge of inconsistent read depth. Variation in read depth without normalization (A), and consistency in read depth with normalization (B).
How DNA normalization works
Each library prep used in a multiplexed DNA sequencing run is unique in terms of both content and concentration. The final concentration depends on the efficiency of your DNA extraction protocol, and quality and quantity of starting material. Evening out these libraries through normalization helps produce consistent and reliable NGS data.
There are opportunities for normalization at several stages of a multiplexed sequencing workflow. You might normalize the concentration of input DNA, size distribution of library fragments, and concentration of library prep before pooling.
Checking the concentration of library preps can have a direct effect on clustering efficiency, clonal amplification, and read uniformity across the pooled libraries. So, standard protocols will often involve quantitatively checking individual library preps and adjusting them to equimolar ratios before pooling. This helps to make sure that all libraries are represented equally on the flow cell.
Methods of quantitating NGS libraries
There are several options for quantitating library preps, varying in ease and accuracy.
- The quickest and most convenient methods (i.e. spectrophotometry-based) tend not to be that accurate.
- The most accurate methods, like quantitative PCR (qPCR), take time and precision, and rely on knowing the average fragment size in each library for dilution calculations (adding more steps to the workflow).
One crucial factor influencing the accuracy of quantitation and subsequent normalization is whether the quantitation method can specifically count adaptor-ligated (i.e. amplifiable) double-stranded DNA (dsDNA) molecules. These are the only molecules that will cluster on the flow cell and contribute to sequencing output.
Illumina’s best practice suggests using fluorometric or qPCR-based quantitation with genomic DNA samples in most cases. Table 1 summarizes the common methods for quantitation.
Table 1. Common approaches to NGS library prep quantitation for normalization.
| Method | Description | Advantages and disadvantages |
| Spectrophotometry | Detects the absorption of UV light by molecules in the sample, with concentration calculated against a standard curve. Estimated purity is based on the ratio of measured absorbance at 260 and 280 nm. | Advantages:
Disadvantages:
|
| Electrophoresis | Estimates fragment sizes through capillary electrophoresis, and concentration through intercalating dyes. | Advantages:
Disadvantages:
|
| Fluorometry | Uses dsDNA-specific intercalating fluorescent dyes for assessing the concentration of nucleic acids against a standard curve. | Advantages:
Disadvantages:
|
| Quantitative PCR (qPCR) | Probe-based chemistries use adaptor-specific primers with fluorescent dyes and quenchers to quantitate library preps against standard curves. Digital droplet qPCR is a variation that can provide absolute quantitation without reference samples. | Advantages:
Disadvantages:
|
qPCR provides the ultimate accuracy in quantitation
It’s interesting that no single method provides all the data you need with enough accuracy for normalization. Though fluorometry and qPCR enable the most accurate quantitation, neither can estimate average fragment size. So, it’s often still necessary to check this by electrophoresis.
Of these two most accurate methods, only qPCR can specifically target the adaptor-ligated molecules. It uses primers complementary to the adaptor sequences. Quantitating only these viable sequencing templates gives you the best chance at normalizing your libraries accurately.
Adaptor ligation efficiency can vary between individual samples and batches. It’s reliant on enzymatic reactions that could be affected by impurities and differences in the quality of starting material. So, quantitating with no specificity for adaptor-ligated molecules (fluorometry) means you’re more likely to overestimate the sequencing-competent library concentration and over-dilute.
Having said that, if your starting material is of high and consistent quality, and the end repair/adaptor ligation step of your library prep workflow is efficient, fluorometry can be a cheaper, faster, and nearly as accurate an option.
If you’re looking for the ultimate accuracy in quantitation though, qPCR is the way to go.
Magnetic beads-based normalization as an alternative
What if you didn’t need to go through the trouble of quantitating your libraries at all?
It’s increasingly common to find magnetic beads popping up in NGS sample prep workflows. For example, they are already being used for size selection—another challenge in NGS sample prep—providing a reliable and established way to safely handle nucleic acids.
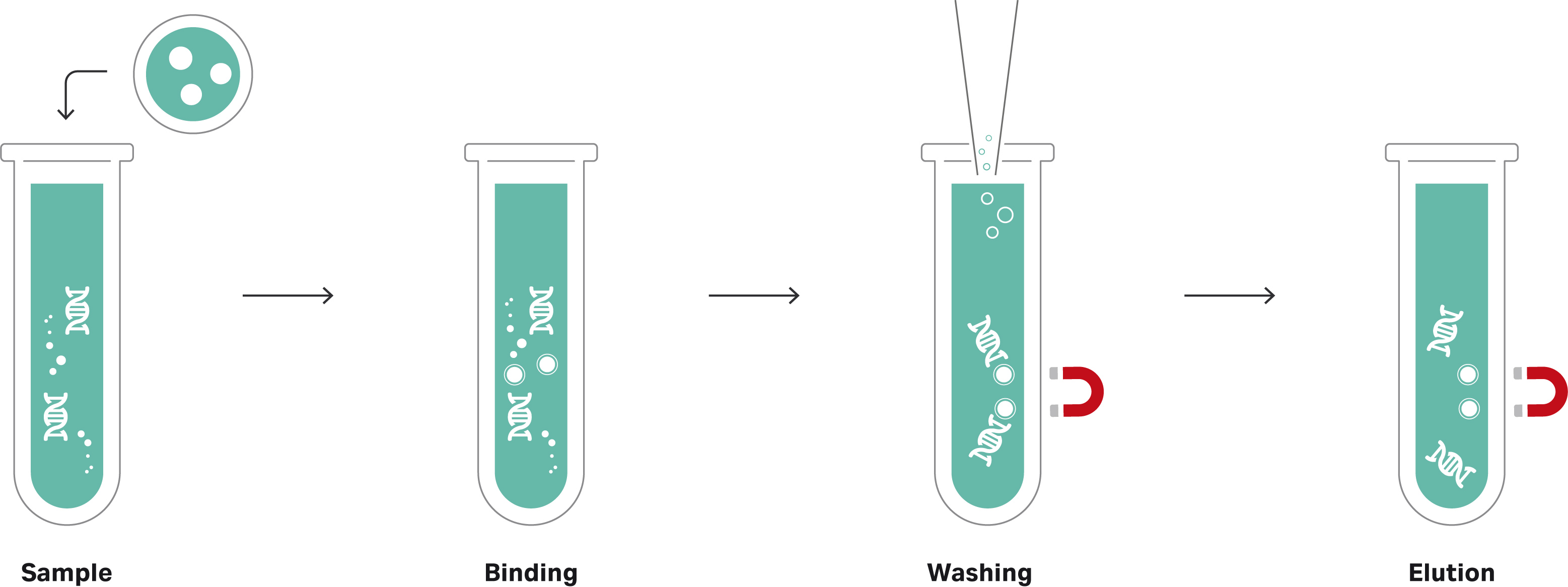
The idea behind magnetic bead-based normalization is that a given volume of beads can bind a consistent quantity of nucleic acid molecules. That is, if there are enough molecules in each library to saturate the beads, an essentially equimolar quantity of library fragments will bind and be retained from each sample (Fig 2). All unbound molecules are then washed away so that each library is represented by just the bead-bound molecules.
Fig 2a. Principle of magnetic bead-based normalization of DNA libraries.
Fig 2b. Principle of magnetic bead-based normalization of DNA libraries with silica core.
There are several coating options available to suit any given application: carboxyl- and silica-coated magnetic beads for generic, non-specific binding based on buffer conditions; oligo(dT)-coated beads for binding mRNA; and streptavidin-coated beads for binding biotinylated samples.
This approach is reasonably straightforward and studies in recent years have indicated that bead-based normalization produces more consistent read depth than several existing quantitation-based methods. Illumina has exploited this approach for normalization, modifying its transposon-based ‘tagmentation’ system for NGS library prep to use magnetic beads.
The bead-based approach, however, can be wasteful: the number of molecules in each library needs to equal or exceed the binding capacity of the beads, with the excess discarded. If your sample is precious or in short supply, it might be worth taking the extra time for qPCR-based quantitation.
Best practice for selecting a normalization method
Use fluorometry for library normalization when:
- Your samples are of good quality
- Your sample prep workflow has a history of producing consistent concentrations
- Some variation in quantitation, and so read depth, is acceptable
Use qPCR for library normalization when:
- Your samples are from varied sources, are precious, or in limited supply
- You need the ultimate accuracy for normalization
- It’s essential that you achieve a minimum target read depth
Use magnetic beads for library normalization when:
- Your samples are in plentiful supply but might vary in quality
- Your library prep yields are usually high (at least 10–15 nM, according to Illumina best practice)
- You have many samples and need to minimize time spent quantitating
At Cytiva, our genomics experts aim to support you in all aspects of your NGS workflows. Read our other genomics blogs for news, tips, and insights. To find out more about optimizing your NGS library preps, or for support in any other aspect of your workflow, contact Cytiva Scientific Support or your local Cytiva representative.