Is PCR always the most appropriate choice for DNA amplification? Find out how Phi29-based isothermal amplification can complement PCR in molecular biology workflows, simplifying NGS and single-cell sequencing applications.
Understanding genetic diseases, rapidly diagnosing infections, and performing forensic investigations requires sophisticated workflows, such as next generation sequencing (NGS), that rely on high quality DNA. But, obtaining enough DNA from some types of sample, such as single cells, is a challenge.
What do you do when you have limited source material?
Sample preparation protocols often account for this challenge with an optional amplification step, depending on the quality and quantity of starting material. This step can mean the difference between a successful and unsuccessful sequencing run. But not all amplification methods are the same.
In this blog, I will review and compare the features and applications of polymerase chain reaction (PCR), the old workhorse of the molecular biology lab, and multiple displacement amplification, based on Phi29 DNA polymerase, an alternative approach that is gaining traction.
What is PCR?
PCR has been such a staple of the modern laboratory that it does not need much of an introduction. Developed by American biochemist, Kary Mullis, in 1983, the amplification method involves three steps, usually repeated for 20 to 30 cycles, or as is optimal for the application:
- Melting the DNA template at a high temperature (e.g. 95°C).
- Annealing oligonucleotide primers at a lower, optimized temperature (e.g. 56°C).
- Extension of primers by a DNA polymerase, often Taq DNA polymerase or a variation at 72°C.
The method has become embedded in many laboratory protocols due to its simplicity, specificity, and predictability. It is a critical component of basic and applied research, forensics, diagnostics, and has even found a place in NGS workflows.
An NGS protocol might, for example, call on high-fidelity polymerases—those discovered or engineered to have improved discrimination of nucleotides and proofreading capabilities—for whole genome amplification.
If performed on starting material, this step can bypass the need for later library fragmentation by generating amplicons of appropriate size. Or, if performed after fragmentation, this step can incorporate index barcodes or other functional elements at the same time.
In targeted sequencing, PCR provides a simple method of target enrichment, and there are numerous commercial kits on the market designed for this purpose.
Limitations of PCR
While it is no surprise then that PCR became the workhorse of the lab, it has limitations.
The method usually requires a known target template sequence for priming, and the primers often require optimization, especially when multiplexing with multiple primer sets. The reaction mixture also contains dNTPs, magnesium ions, and other PCR reagents that require optimization.
There are bioinformatic tools that can assist in designing primers and optimizing temperature cycling conditions, but a process of trial and error is inevitable.
Numerous efforts over the decades have improved on the original Taq DNA polymerase or identified alternatives, but even these higher fidelity polymerases have limitations.
In workflows requiring faithful amplification of template, such as NGS, minimizing the risk of introducing unwanted errors in analysis requires both high-fidelity polymerases and limiting the number of cycles. Applications with limited source material, as in single cell sequencing, exacerbate this challenge.
In some situations, another approach—Phi29-based amplification—can provide a simpler and complementary alternative to PCR.
What is Phi29 amplification?
Identified in the 1980s, Phi29 (Φ29) DNA polymerase comes from the bacteriophage Φ29. It is now finding increasing use in molecular biology for multiple displacement amplification (MDA) procedures, and has several features that make it well suited to this application:
- Isothermal polymerase activity
- Strong strand displacement
- 3’-5’ exonuclease proofreading
The combination of these features makes Phi29 an attractive option for sensitive applications where MDA might be better suited than PCR.
Phi29 exhibits a 100-fold higher fidelity compared to standard Taq polymerase, thanks to its proofreading ability, placing it at a similar level to other high-fidelity enzymes.
However, key is the isothermal amplification. The strong strand displacement removes the need for temperature cycling and optimization, resulting in a simpler process than PCR: one temperature, one ‘cycle’, no optimization necessary.
This mechanism enables the continuous replication of template, leading to exponential amplification and two key applications: rolling circle amplification (RCA) and whole genome amplification (WGA).
Rolling circle amplification with Phi29
From nanograms of initial DNA sample, you can use Phi29-based MDA to rapidly produce microgram yields (or more) of high-quality DNA ready for direct use in a range of applications.
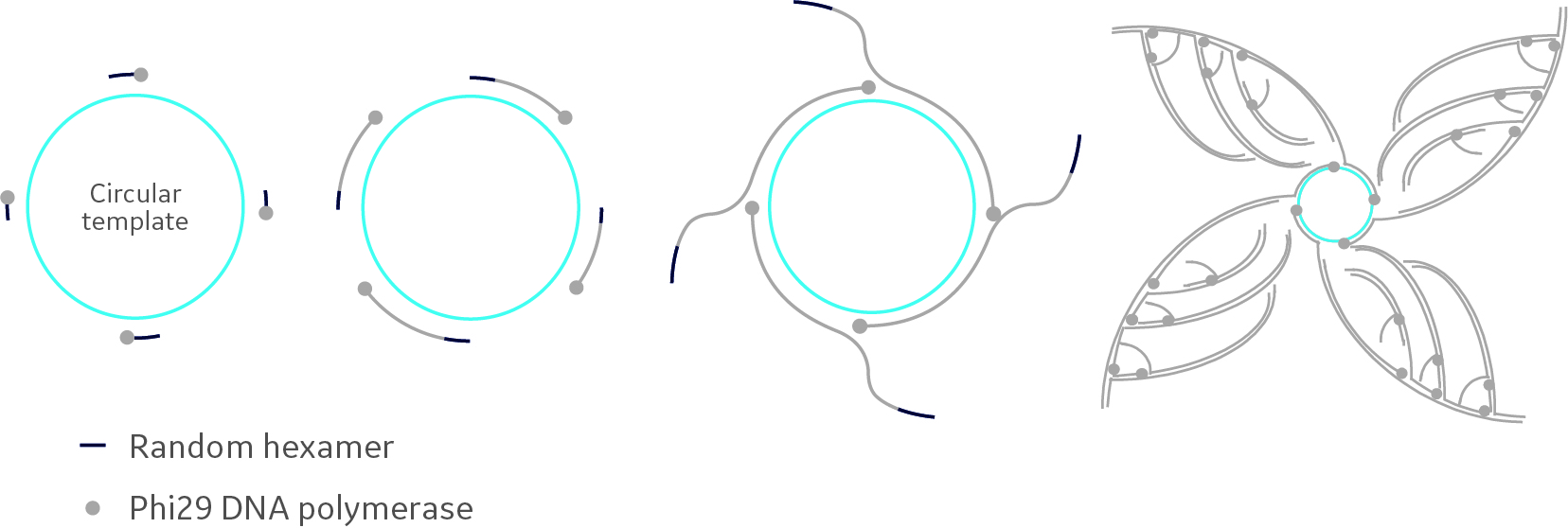
In RCA, Phi29 acts on a circular DNA template, such as a plasmid vector, using random hexamer primers. Kept at a constant 30°C, the Phi29 DNA polymerase produces long ssDNA strands containing hundreds to thousands of tandem repeats of the initial template. These strands can then act as templates for further priming, branching into more copies and an exponential amplification of the original circular template (Fig 1).
Fig 1. Principle of rolling circle amplification. Random hexamer primers anneal to the circular template DNA at multiple sites. Phi29 DNA polymerase extends each of these primers. Strand displacement occurs when the polymerase reaches a downstream-extended primer. Subsequent priming leads to an exponential isothermal amplification.
This mechanism makes RCA well suited for cell-free cloning. In a few hours, RCA can generate micrograms of vector from colonies or glycerol stocks directly, removing the need to culture overnight. The same principle applies to any circularized template.
Whole genome amplification with Phi29
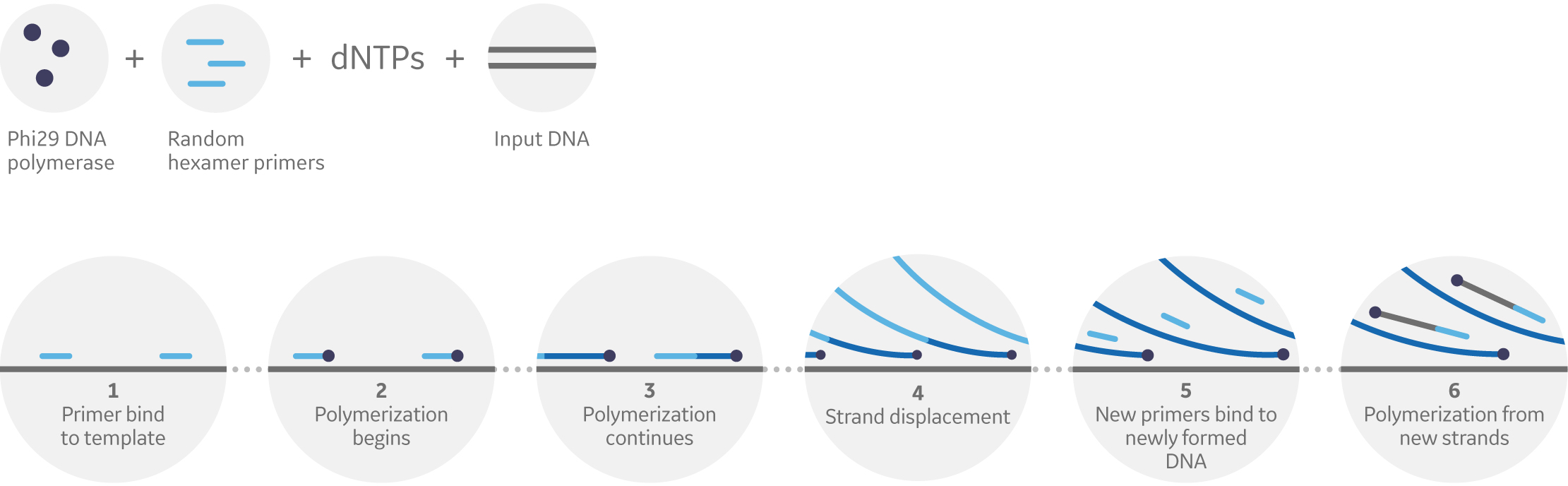
The WGA application of Phi29 works similarly to RCA but increases the scale dramatically. Through multiple displacement, the method binds primers to newly-formed DNA while polymerization is still ongoing across the entire genome. The new strands continue to branch, leading to a rapid and exponential amplification (Fig 2).
Fig 2. Principle of Phi29 DNA polymerase-based multiple displacement amplification. Template DNA is primed at multiple locations with random hexamers. Phi29 DNA polymerase then extends primers, replicating template and displacing any downstream extended primers. Strand displacement and subsequent priming leads to an exponential increase of new template for isothermal amplification.
With the ability to amplify picograms of starting material using optimized formulations, Phi29-based amplification enables you to take a single cell and generate more than enough DNA for reliable NGS. Indeed, you can even use the amplified DNA directly for sequencing or put it through library construction without the need for more purification (unlike PCR-based amplification), further simplifying the workflow.
Ultimately, in either application, Phi29-based multiple displacement amplification means you can reduce hands-on time in otherwise complex workflows without compromising sequencing success or read length.
Applications of PCR and MDA
While isothermal amplification using Phi29 has several benefits over traditional PCR, it is not a replacement for the old workhorse. PCR remains a key component of many molecular biology workflows, such as sequencing, genotyping, and site-directed mutagenesis.
For situations with amplicons within the reliable limit of high-fidelity polymerases (usually less than 10 kbp) and known sequences (or at least primer binding sites), PCR remains the first choice.
In other situations, WGA for single-cell sequencing for example, you have more options. There are PCR methodologies for WGA, but they are relatively complex, potentially less robust, and require more optimization than MDA.
Using Phi29-based MDA could provide a faithful and uniform amplification of the original genome with fewer variables, larger amplicons, and less effort than PCR. But during library prep, you are again likely to return to PCR for steps like end-repair and adding functional elements to your fragments.
Beyond sequencing, however, the features of Phi29 DNA polymerase also lend themselves well to several other applications (Table 1). For example, the high fidelity, and therefore low rates of false positives and negatives, make Phi29 well suited for identifying single nucleotide polymorphisms (SNPs) and other mutations.
Table 1: Comparison of PCR and MDA approaches
| Amplification method | PCR | MDA |
|---|---|---|
| Key features |
|
|
| Application examples |
|
|
As workflow for MDA is also simpler than PCR, requiring no thermal cycling, commercial kits offer straightforward one-tube, one-temperature formats that facilitate automation and high-throughput sample amplification.
Availability of commercial Phi29 amplification kits
We offer several Phi29 amplification kits, TempliPhi DNA amplification kits for RCA and GenomiPhi DNA amplification kits for WGA.
Designed to eliminate overnight culture steps for plasmid/fosmid glycerol stocks and colonies, the amplified DNA from our TempliPhi kits is ready for direct use in sequencing or your chosen application, streamlining the entire process and reducing hands-on-time.
Our GenomiPhi kits come in two formats, traditional liquid and Ready-To-Go. The Ready-To-Go kits contain pre-dispensed, room-temperature stable reaction mixes. Simply add your starting material and let the reaction run.
The manufacturing processes for these kits involve UV and enzymatic reagent cleanup that help make sure they are free of detectable DNA contamination and permit sensitivity of DNA amplification down to the 1 femtogram, amplified to several micrograms in a just a few hours.