Home in on the nature of interactions
Interactions between biomolecules are the key drivers of any biological process. For example, protein-protein interactions ensure signal transduction across membranes via G protein-coupled receptors (GPCRs) and maintain the structure of complexes by chaperones. These types of interactions also allow enzymatic modification of proteins via post-translational phosphorylation. Hundreds of thousands of interactions have been identified and collected in databases, for example IntAct. Such a database allows the interactions to be assembled into pathways and studied further.
Did you know you can simplify research on protein-protein interactions using Biacore™ SPR systems? Check out this peer-reviewed article for inspiration:
Perform kinetics studies to link structure and function
The cell is a dynamic system. So, identifying interactions is just the first step in interaction analysis which often leads to an investigation of kinetics to answer the following questions:
- How fast do molecules bind (association)?
- How fast do complexes fall apart (dissociation)?
Kinetics determine whether a complex forms or dissociates within a given time span. Association and dissociation measurements can also be used to determine how much complex is formed at equilibrium (the steady state where association balances dissociation). This is the affinity of the interaction. Studying kinetics allows linking between structure and function and provides a greater understanding of biological mechanisms.
Kinetics studies are simple to perform using Biacore™ systems (Fig 1). Read these research articles to learn how your peers are doing it:
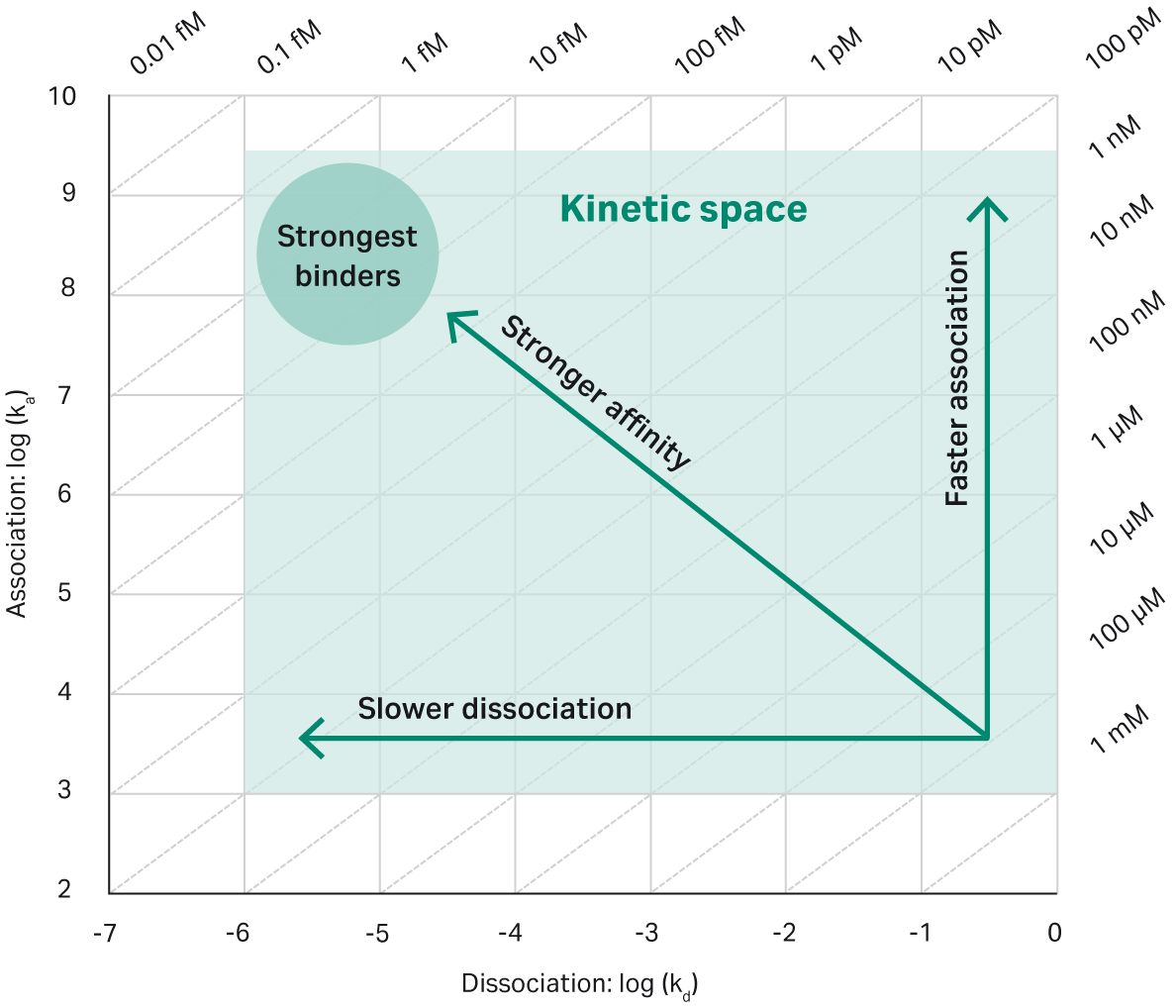
Fig 1. Kinetic measurements over a broad range, from the fastest on-rates to the slowest off-rates. Even interactions at the extremes of kinetic behavior (e.g., with very slow off-rates) can be detected and differentiated with confidence.
Learn how proteins work by looking at their domains
The structure of a biomolecule is usually critical to its function. The structure may be static. But usually it’s dynamic and influences, or is influenced by, interactions with other biomolecules. Very often, as in the case of enzymes, changes in structure are instrumental to function. Protein structure may be determined by sequencing. Then, biophysical methods such as X-ray crystallography and nuclear magnetic resonance (NMR) are applied to determine the three-dimensional structure as the protein interacts with cofactors. Understanding the roles conformational changes and structure play in interactions requires other methods. These methods include biochemical assays and biophysical techniques, such as SPR-based assays.
Read how these researchers are exploring protein domains using Biacore™ systems:
Get answers using surface plasmon resonance
Because surface plasmon resonance provides so much rich data, it can help answer key questions about the nature of binding. It also allows you to fully understand binding events between almost any type of biologically relevant interactants. Here are just some of the questions you can delve into:
- Do the interactants bind to each other?
- How specific is the interaction?
- How strong is the binding (affinity)?
- How fast are the on- and off-rates (kinetics)?
- What are the effects of temperature on binding (thermodynamics)?
- How much interactant is in the sample (concentration detected via specific binding partner)?
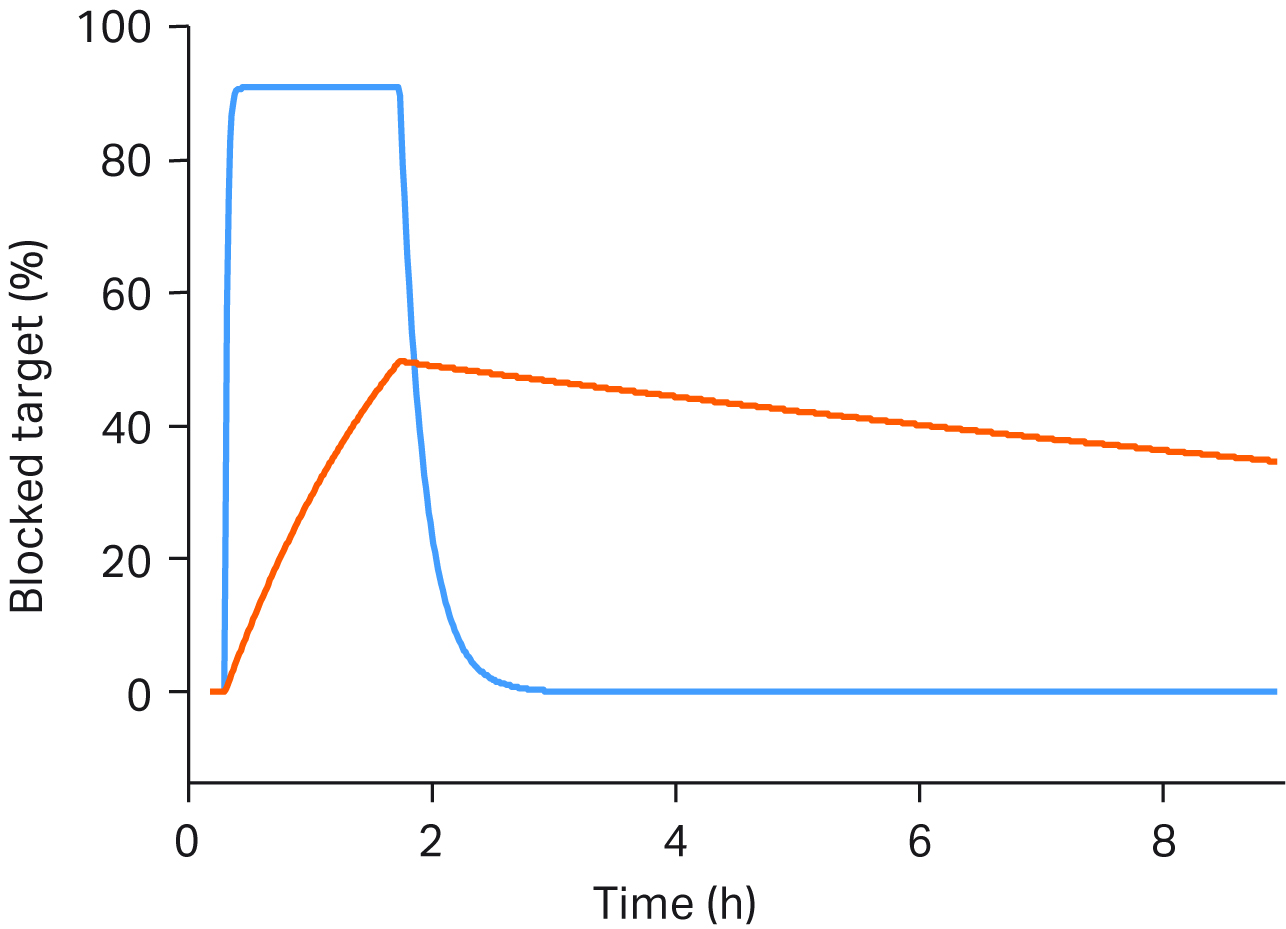
If you use end-point techniques today, you’re only getting basic information such as overall binding strength (affinity). Because affinity depends on the ratio of on-and off rates, interactions with the same affinity can have very different kinetic properties. Real-time, label-free interaction analysis with Biacore™ SPR systems can provide the key data to discriminate these crucial differences even for interactions with challenging targets (Fig 2).
Fig 2. Blue sensorgram illustrates rapid kinetics: frequent administration of a low dose to occupy the target. Orange sensorgram illustrates slower kinetics: administration of a high dose to occupy the target for a long time.
Biacore™ technology is cited in 60 000 peer-reviewed scientific articles – and counting. Who uses it?
- Academic research institutes and universities worldwide, to define and characterize key molecules and understand the functional interactions
- Pharmaceutical and biotechnology companies around the world, for screening, characterization, and quality control of therapeutics
- Diagnostic and research tool companies, to provide characterized antibodies and reagents
Access even more references here:
Choose SPR and Biacore™ systems for your research, if you value:
- Robustness and reproducibility. In a market survey of more than 100 Biacore™ system users, over 90% of responders are satisfied with the robustness and reproducibility of our systems. Over 50% are extremely satisfied.
- High sensitivity and a wide kinetics range. This means that you can run a wide range of assay types, applications, and a broad range of molecules and samples.
- Extensive support. You have easy access to assay set-up tools with methods, application procedures, and self-learning. Or you can attend live training courses.
- In-house application expertise. You get access to our experienced application scientists who develop new applications and new methodology to meet your needs.
- Ease of use. You can get high-quality data even with limited know-how. Over 60% of surveyed users are extremely satisfied with our ease of use, and40% are satisfied.
- Innovation. We led the way with supervised data evaluation support using machine learning. This innovation saves time spent on data interpretation and minimizes differences between users.
- Global and local presence: Our application scientists and system maintenance engineers are at your service globally as well as locally.
- High-quality data. Designed to provide high-quality data, our microfluidics provide precise control of sample volumes, without diffusion between sample and buffer while in the fluidic channels. This is true for pure samples as well as complex cell cultures, serum, or plasma samples.
- Affordable. Re-useable sensor chip surfaces allow multiple samples to be run over the same flow cell giving you lower cost per sample. Cytiva offers different Biacore™ system models with up to 16 flow cells.
- Expertise. Over 90% of survey responders believe we have expertise and provide innovative solutions in the label-free interaction analysis market with over 30 years ofexperience. The SPR technology is included in EMEA, US, and Japan pharmacopoeia.