Controlled alkaline lysis of E. coli cells using single-use static inline mixer to release plasmid DNA (pDNA) for further purification and use in biopharmaceutical manufacturing
Plasmid DNA (pDNA) is a prerequisite in the growing biopharmaceutical field of mRNA and in gene therapy manufacturing. Plasmids used in BioProcess™ applications are commonly in the range of 5 to 20 kilobase pairs (kbp). Here we present a controlled scaled-up process using single-use equipment and consumables for an E. coli cell line with 6.3 kbp model plasmid.
Individual lysis process parameters can be adjusted for optimization with the goal of maintaining pDNA quality and yield. We also show how the lysis and clarification process fit into the pDNA workflow and can be used in scaled-up pDNA processes from 50 to 200 L.
Previously published material around pDNA production purification and analysis from Cytiva is available in the following application notes:
- A scalable single-use two-step pDNA purification process
- Three-step supercoiled plasmid DNA purification
Introduction
pDNA is an important genetic engineering tool used to clone and amplify or express genes for biotechnology applications. pDNA of good manufacturing practice (GMP) grade has many applications including DNA vaccines and gene therapy, with the production of viral vectors and mRNA being dependent on the production of pDNA (1).
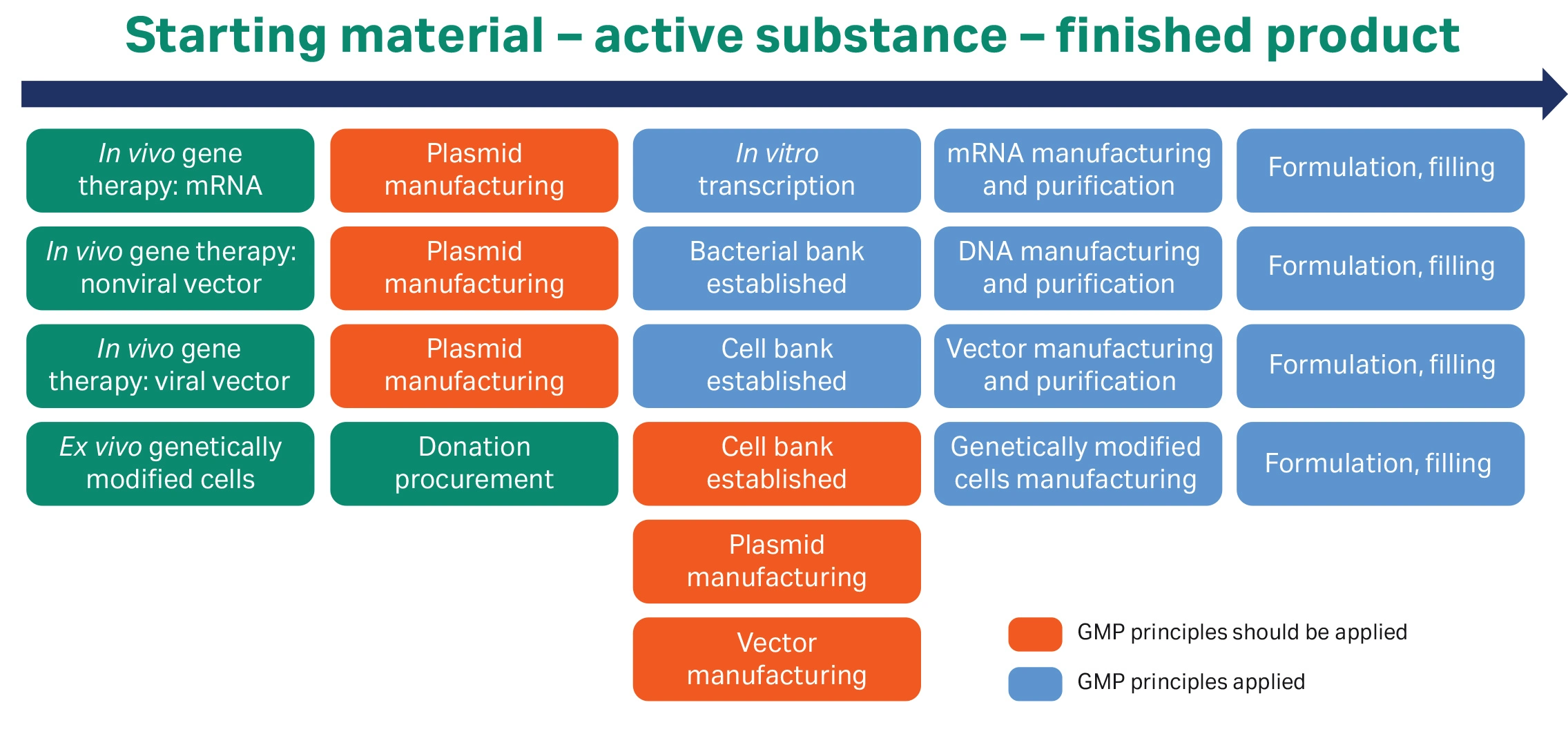
With the increasing use of plasmid vectors in different therapeutic areas, the focus on manufacturability, as well as regulatory requirements for these processes, have increased. Manufacturers of pDNA must comply with GMP principles even when plasmids are not themselves therapeutic (Fig 1 below).
Fig 1. Overview of where GMP principles apply for pDNA manufacturing. Adapted from reference 2.
Plasmids are amplified through a microbial system in a fermentation process followed by a series of purification steps to reach the purity and quality goals. A critical step in the purification process is alkaline lysis, where concentrated cells are suspended at neutral pH and lysed by adding an alkaline solution containing detergent. The lysate is then neutralized, resulting in the immediate formation of a white precipitate or flocculate consisting mainly of cell debris, denatured proteins, and genomic DNA.
The lysis is often performed in batch mode and removal of the flocculate/precipitate is often a centrifugation operation at lab scale. However, the batch mode of the lysis process is difficult to close and scale when fermentation volumes increase.
An additional challenge when performing lysis in batch mode is achieving optimized mixing while minimizing shear during the brief lysis period, which typically lasts only a few minutes at alkaline pH. Prolonged lysis time risks the degradation and denaturation of supercoiled plasmids (SC), while high shear forces risk fragmenting genomic DNA. Both issues can complicate the purification process and reduce yield and quality.
While batch lysis using mixers is available and performed at small scale, control of the lysis process while maintaining appropriate mixing and lysis time parameters can be challenging as cell mass and volumes increase.
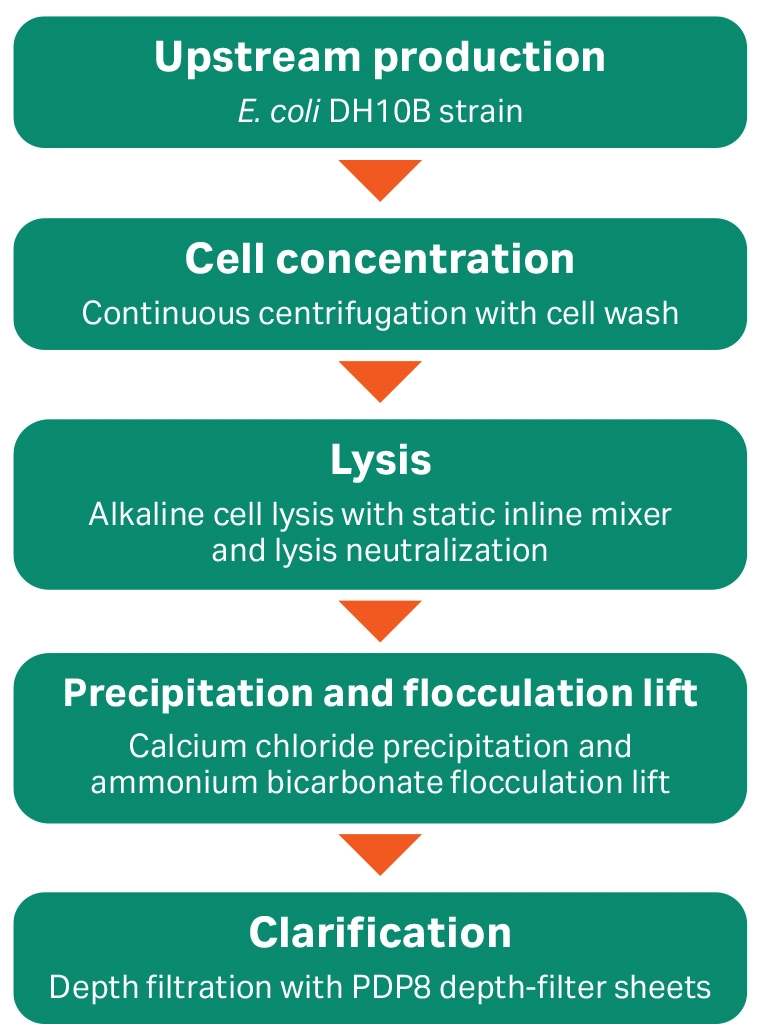
Our main objective of this work is to incorporate a controlled and scalable process for E. coli lysis with single-use equipment and consumables. The pDNA workflow typically includes the lysis step between the cell concentration and clarification depth filtration process steps in the midstream process (Fig 2).
Fig 2. An overview of pDNA production through the midstream process. Lysis was performed directly from diluted cell suspension feed using the single-use static inline mixer.
The alkaline lysis process with the static inline mixer allows for optimized mixing and reduced shear to avoid fragmentation of genomic DNA and denaturation and degradation of SC plasmid. The single-use components allow easy setup, reduce the need for post-use cleaning, and ensure a closed system for the cell concentration and lysis process.
This application note shows the performance of a static inline mixer for alkaline cell lysis and pDNA release from cell suspension feed material. We performed the work at the 50 and 200 L fermenter scale using a single-use centrifuge for cell concentration, followed by a full pDNA midstream process including alkaline lysis operation, calcium chloride precipitation of RNA, clarification through flocculate lift (2), and depth filtration.
Materials and methods are described in detail at the end of the document.
Results and discussion
We produced the pDNA plasmid starting material for two runs in Xcellerex™ XDR-50 MO and XDR-200 dual-purpose (DP) fermenters. E. coli cells were concentrated, washed, and buffer exchanged using a continuous single-use centrifuge where the plasmid-containing cells were retained (heavy phase). The fermenter medium (light phase) was set to waste.
The cells in the heavy phase were retained in a circulation tank, recirculated in the centrifuge, and washed with cell suspension buffer (50 mM Tris, 50 mM glucose, 10 mM EDTA, pH 7.5), while keeping the volume constant, equivalent of > two buffer exchanges.
We determined the wet cell weight (WCW) of the E. coli cells and diluted to a final concentration of 15 L/kg of cell suspension buffer to WCW. For the 50 and 200 L fermenter runs, the recirculation vessel and dilution vessels were single-use Allegro™ 50 L and Xcellerex™ XDUO-200 single-use mixers, respectively. The starting weights of the lysis material were 50.7 and 100.0 kg with processed wet cell weights of 3.44 and 6.66 kg for the two experiments (for the 200 L run only 50% of the WCW from the cell concentration step was further processed through the lysis).
The lysis process employed single-use components throughout, including the static inline mixer assembly, mixer bags, and ReadyToProcess™ tubing assemblies. The ½” static inline mixer is a 19 cm long, 12.7 mm o.d., helical mixer. The single-use components, peristaltic pumps, and pressure monitor were organized on a ReadyKart™ mobile processing station.
The cell suspension feed material was mixed at 30 rpm in the Allegro™ 50 L mixer through the process until reaching the minimum 2 L agitation volume to keep it heterogeneous through the lysis process. The static inline mixer flow rate was controlled at a total flow rate of 500 mL/min through the mixer (per element if multiple elements are used Fig 3B). Half of the flow came from the cell suspension and half from the lysis buffer (0.2 M NaOH, 1% SDS) giving 1:1 proportion of the two solutions.
The 500 mL/min flow rate corresponds to a flow velocity of 0.067 m/s. The target was 1 min of lysis time and the length of the 1/2” i.d. tubing, post static inline mixer was scaled to match this time (4 m for the 50 L run and 7.8 m for the 200 L run). Tubing was coiled in ~ 0.3 m (1 ft) diameter loops before the introduction of the neutralization buffer (3 M potassium acetate, 2.07 M acetic acid at 2°C to 8°C) at a Y junction followed by collection into single-use mixers (XDUO 200 L mixer for the 50 L run and XDUO 500 L mixer for the 200 L run).
Figure 3 shows the setups of the runs.
Fig 3. Setup for the pDNA lysis process for the 50 L fermenter scale (A) and 200 L fermenter scale (B) using two static inline mixer elements to optimize process time. (C) Post-lysis operations with calcium chloride and ammonium bicarbonate.
The inlet pressure of the static inline mixer was consistent through the entire runs at 0.25 to 0.35 bar (Fig 4) and showed no sign of clogging during the runs. Processing time for each of the runs was ~ 3 h.
Fig 4. Pressure before static inline mixer(s) during lysis runs.
The final neutralized lysate weighed 147.7 and 311.1 kg, collected into XDUO-200 or XDUO-500 mixers for the 50 and 200 L runs, respectively. The lysis process yielded > 90% of pDNA released from the lysis process based on pDNA titers in the neutralized lysate product as compared to titer from fermenter samples processed with pDNA miniprep kits (Table 1).
Product quality data (Table 2) post-lysis shows that the static inline mixer lysis did not affect the pDNA supercoiled/open circular (SC/OC) ratio and product impurities between runs.
Table 1. End of fermentation through lysis process data. End of fermentation concentration performed via pDNA miniprep kits and spectrophotometry. Continuous centrifugation process step yield was calculated based on the percentage of cell mass at the end of fermentation compared to the continuous centrifugation amount1
| Process sample | Fraction weight (kg) | pDNA conc. (mg/mL) | pDNA amount (g) | Step yield (%) | Overall process yield (%) | |||||
| Xcellerex™ XDR fermenter | XDR-50 | XDR-200 | XDR-50 | XDR-200 | XDR-50 | XDR-200 | XDR-50 | XDR-200 | XDR-50 | XDR-200 |
| End of fermentation1 | 37.0 | 200.0 | 0.17 | 0.07 | 6.29 | 14.2 | N/A | N/A | N/A | N/A |
| Continuous Centrifugation2 | 36.18 | 41.68 | N/A | N/A | 5.47 | 10.3 | 87% | 73% | 87% | 73% |
| Neutralized lysate3 | 147.7 | 311.1 | 0.034 | 0.018 | 5.02 | 5.60 | 92% | 109% | 80% | 79% |
1End of fermentation sample pDNA concentration analysis was performed using a NanoDrop microvolume spectrophotometer on batch lysis material purified using DNA miniprep kit (see Materials and methods for additional information).
2Continuous centrifugation pDNA amount data was extrapolated based on process step yield and WCW retained during continuous centrifugation.
3The 200 L run only forward processed 50% of the material from the cell concentration through the lysis process step. The step yield and overall process yield data were calculated accordingly. The neutralized lysate sample pDNA concentration analysis was performed via the Sepharose™ HP chromatography resin method (3) (see Materials and methods for additional information). The 200 L Xcellerex™ XDR fermenter run had a paired yield excursion with continuous centrifugation and neutralized lysate step yields. The overall process yields at the neutralized lysate stage were comparable between the 50 and 200 L runs.
Table 2. Fermenter and lysis analytical data1
| Process sample | E. coli DNA (µg gDNA/mg pDNA) |
E. coli HCP (µg HCP/mg pDNA) |
Endotoxin (E.U./mg pDNA) |
SC pDNA (%) |
||||
| Xcellerex™ XDR fermenter | XDR-50 | XDR-200 | XDR-50 | XDR-200 | XDR-50 | XDR-200 | XDR-50 | XDR-200 |
| End of fermentation1 | N/A | N/A | N/A | N/A | N/A | N/A | 66.6% | 55.8% |
| Neutralized lysate | 126 | 829 | 812 | 490 | 70 340 | 32 350 | 50% | 60% |
1See Materials and methods section for additional details on analytical methods.
We recommended that you perform lysis optimization for specific E. coli cell lines and plasmids to improve product quality and yield. Some key parameters for the lysis process to optimize are:
- Cell suspension dilution
- Lysis and neutralization buffer composition and detergent concentration
- Lysis time/flow rate
Post-lysis, a calcium chloride precipitation, and flocculation lift with ammonium bicarbonate was performed within the Xcellerex™ XDUO mixers. We added 5 M of calcium chloride buffer to precipitate and reduce RNA. We also added 120 g/L of ammonium bicarbonate buffer to bring the precipitated material (precipitated RNA and cell debris) to the top of the mixer bag and separate it from the lysis suspension with the target pDNA. This allows the lysis material to be depth-filtered without loading the precipitated material onto the depth filters, which increases the filtration load capacity. Additions of both buffers occurred through the bottom port for addition close the mixer (Fig 3C).
We performed depth filtration with Stax™ disposable depth filter capsules and PDP8 filter membranes from the bottom outlet of the Xcellerex™ XDUO mixer. Filtration was stopped when the filtration of precipitated material started. Cake filtration with the K900P depth filter and filter aids (Celpure C300 and Harborlite 900S) was tested on the precipitated lysis material at small scale with similar results (data not shown) to flocculate lift material shown in this study for additional clarification options.
Step yields and individual analytics for the precipitation through depth filtration process steps were not quantitated until after tangential flow filtration (TFF) 1 buffer exchange due to chemical incompatibilities with assays.
Conclusions
- Single-use, static inline mixer for E. coli lysis and pDNA release provides a controlled and scalable lysis process with step yields around 90%.
- Processing of a 50 L fermenter batch with one static inline, single-use mixer in ~ 3 h.
- Process steps can be closed operations without the need for cleaning in place (CIP) of equipment.
- Ability to optimize lysis parameters for product quality and process yield targets.
- Multiple single-use mixer sizes available for use (½” and ¼” i.d. static inline mixers) in tubing sets.
- Scalable clarification process (precipitation or flocculation lift and depth filtration) for efficient removal of cell debris and precipitated material after lysis.
CY45846
REFERENCES
- EMA/246400/2021, Questions and answers on the principles of GMP for the manufacturing of starting materials of biological origin used to transfer genetic material for the manufacturing of ATMPs. https://www.ema.europa.eu/en/documents/other/questions-answers-principles-gmp-manufacturing-starting-materials-biological-origin-used-transfer_en.pdf
- Blom H, Bennemo M, Berg M, Lemmens R. Flocculate removal after alkaline lysis in plasmid DNA production. Vaccine. 2010 Dec 10;29(1):6-10. doi: 10.1016/j.vaccine.2010.10.021.
- Bennemo M, Blom H, Emilsson A, Lemmens R. A chromatographic method for determination of supercoiled plasmid DNA concentration in complex solutions. J Chromatogr B Analyt Technol Biomed Life Sci. 2009 Aug 15;877(24):2530-6. doi: 10.1016/j.jchromb.2009.06.037.
MATERIALS AND METHODS
Two experiments were performed, fermentation in a 50 L fermenter and a scaled-up process to a 200 L fermenter. Both experiments will be described in the sections below.
pDNA production
E. coli DH10B strain with integrated target 6.3 kbp plasmid were grown in Terrific Broth (TB) with a glucose-based feed using the Xcellerex™ XDR-50 MO and XDR-200 DP fermenters. Fermentation was performed at 37°C for 16 to 17 h. Fermentation data and equipment are captured in Table 3.
Table 3. Fermentation procedure for 50 and 200 L fermenter scales
| Step | Procedure | Details | 50 L XDR run | 200 L XDR run |
1 |
Fermentation |
Fermentation process duration (h): | 16 | 17 |
| Final fermenter weight (kg): | 37.0 | 200.0 | ||
| Final fermenter OD600: | 52.0 | 42.5 | ||
| Final fermenter, solids (%): | 10.7 | 9.2 | ||
| Fermenter total cell mass (kg): | 4.0 | 18.4 | ||
| Fermenter equipment: | Xcellerex™ XDR-50 MO | Xcellerex™ XDR 200 Dual Purpose |
Cell concentration
We chilled fermenter product to 20°C in the fermenter before starting cell concentration. Centrifugation was performed with an Alfa Laval CultureOne Primo or CultureOne Maxi single-use centrifuge with Allegro™ 50 L mixer or Xcellerex™ XDUO-200 mixer as recirculation vessel, for 50 and 200 L runs, respectively. The centrifuge heavy phase was retained and recirculated back to the recirculation vessel. We maintained a constant weight of the recirculation vessel by adding the cell wash buffer to the top of the vessel for buffer exchange.
The recirculation vessel was buffer exchanged three times for the 50 L run. We buffer exchanged the 200 L run two times, then we collected the heavy phase during the final pass into a 50 L Allegro™ mixer. The high percentage of solids of the heavy-phase collection during the 200 L run at 32.0% may have a higher degree of assay variability due to the higher percentage of solids. At the end of centrifugation, we flushed the centrifuge with cell wash buffer via the centrifuge feed line. The centrifuge light phase was collected as waste and stayed below 30 nephelometric turbidity units (NTU) throughout both runs (data not shown). We measured turbidity using a LaMotte 2020we turbidimeter with offline samples taken from the light phase line. Cell concentration data, buffers, and equipment are described in Table 4.
Table 4. Cell concentration procedure for 50 and 200 L fermenter scales
| Step | Procedure | Details | 50 L XDR-MO run | 200 L XDR-200 DP run |
2 |
Cell concentration |
Centrifugation process duration (h) | 1.9 | 2.6 |
| Buffer exchanges | Three times | Two times (three times during cell suspension dilution) | ||
| Post centrifugation weight (kg) | 32.20 | 41.68 | ||
| Post-centrifugation solids (%) | 9.5 | 32.0* | ||
| Post-centrifugation total cell mass (kg) | 3.44 | 13.3 | ||
| Cell wash buffer | 50 mM Tris, 10 mM EDTA, 50 mM glucose, pH 7.5 | |||
| Single-use centrifuge equipment | Alfa Laval CultureOne Primo | Alfa Laval CultureOne Maxi | ||
| Recirculation vessel | Allegro™ 50 L mixer | Xcellerex™ XDUO 200 mixer | ||
| Cell-wash buffer transfer pump | Watson Marlow 520 series peristaltic pump | Watson Marlow 520 series peristaltic pump | ||
* The heavy phase collection during the 200 L run at 32.0% and low yield of step may be caused by the high wet cell weight test variability at higher percentage of solids.
Lysis with static inline, single-use mixer
We diluted the post-centrifugation cell suspension material with the cell suspension buffer (50 mM Tris-HCl, 10 mM EDTA, 50 mM glucose, pH 7.5) to a target of 15 L/kg of cell suspension buffer to total cell mass in the Allegro™ 50 L mixer or Xcellerex™ XDUO-200 mixer. The 50 L run was set up using ReadyToProcess™ single tubing assemblies with ReadyMate™ connections with the inline static mixer. The cell suspension was continually mixed until the minimum agitation volume of 2 L was reached on the Allegro™ 50 L mixer.
The 200 L run utilized two static inline mixers run in parallel, which were set up at identical heights and lengths of inlet and outlet tubing from static mixers. We tested flow rates through the static mixers and within 5% of flow rate through each mixer before combining flows through the ½” i.d. lysis tubing. The 200 L run used both an initial cell suspension mixer (Xcellerex™ XDUO-200) and a surge vessel (Allegro™ 50 L mixer) to maintain mixing through the process step.
We initially filled the Allegro™ 50 L mixer to 20 kg with cell suspension feed and when the minimum mixing volume of the Xcellerex™ XDUO-200 mixer was reached (44 L), the remaining cell suspension was pumped into the surge tank to continue mixing to the minimum agitation volume of 2 L.
Peristaltic pumps for the cell suspension, lysis buffer, and neutralization buffer were standardized at the set flow rate before runs. We primed all lines up to static inline mixer or Y junctions before starting the lysis process step. The cell suspension and lysis buffer pumps were started at the same time and the neutralization buffer pump was started when the lysis product reached the Y junction. The cell suspension feed material was mixed at ~ 30 to 50 rpm in the Allegro™ 50 L mixer and Xcellerex™ XDUO-200 mixer until the minimum agitation volume of each was reached to keep the cell suspension heterogeneous through the lysis process.
The neutralization buffer was chilled between 2°C and 8°C before use. For the 200 L run, we processed only 50% of the centrifuged cell concentration material through the lysis step. The static inline mixer lysis data, buffers, and equipment are described in Table 5.
Table 5. Lysis procedure for 50 and 200 L fermenter scales
| Step | Procedure | Details | 50 L XDR run | 200 L XDR run |
3 |
Lysis with static inline mixer |
Lysis process duration (h) | 3.2 | 3.2 |
| Total cell mass processed (kg) | 3.44 | 6.66 (50% forward processed) |
||
| Cell suspension dilution (L of buffer/kg of cell mass) | 15 | ~ 15 | ||
| Weight of cell suspension feed (kg): | 50.7 | 100.0 | ||
| Number of static inline mixers used | 1 | 2 (in parallel) |
||
| Cell suspension, lysis buffer, and neutralization buffer pump flow rates (L/min) | 0.250 | 0.500 | ||
| Individual static inline mixer flow rate (L/min) | 0.500 | ~ 0.500 | ||
| Individual static inline mixer velocity (m/s) | 0.066 | 0.066 | ||
| Lysis time post-static inline mixer | 1 min (4 m of ½” i.d. tubing) |
1 min (7.8 m of ½” i.d. tubing) |
||
| Cell suspension buffer | 50 mM Tris, 10 mM EDTA, 50 mM glucose, pH 7.5 | |||
| Lysis buffer | 0.2 M NaOH, 1% SDS | |||
| Neutralization buffer | 3 M potassium acetate, 2.07 M acetic acid | |||
| Cell suspension, lysis buffer, and neutralization buffer pumps | Watson Marlow 520 series peristaltic pump | Watson Marlow 520 series peristaltic pump | ||
| Cell suspension surge tank transfer pump | N/A | Watson Marlow 620 series peristaltic pump | ||
| Neutralized lysis collection vessel: | Xcellerex™ XDUO 200 mixer | Xcellerex™ XDUO 500 mixer | ||
| Final neutralized lysate weight (kg): | 147.7 | 311.1 | ||
Neutralized lysate precipitation, flocculation lift, and clarification depth filtration
The neutralized lysate material was mixed in the Xcellerex™ XDUO mixer for 2 to 5 min before further processing. Setup of the XDUO mixer bag was completed by attaching single-use tubing sets to the bottom tubing port for precipitation and flocculation lift additions then depth filtration and an air filter were attached to the top port of the bag.
We added 5 M of calcium chloride (CaCl2) through the XDUO mixer bag bottom port while mixing at a slow speed to a final concentration of 1 M CalCl2. Mixing was continued for an additional 2 to 5 min post-addition then stopped and held for ≥ 1 h to allow precipitate to form.
We added 120 g/L of ammonium bicarbonate flocculation lift buffer through the XDUO mixer bag bottom port while mixing to a final concentration of 8 g/L ammonium bicarbonate. Off-gassing during the flocculation lift addition brings the precipitated material to the top of the bag and is held there with excess gas exiting through the air filter on a top port. Flocculated material was held overnight without issue before starting the clarification depth filtration of flocculated lysate material.
We performed clarification depth filtration with Stax™ capsule, HP-Series, and PDP8 filter sheets. We used Stax PDP8 depth filters of 1 and 2 m2 at a filter loading of 165 L/m2 and 187 L/m2, for the 50 L and 200 L runs, respectively. Watson Marlow 620 peristaltic pump with associated tubing sets and SciLog pressure sensors were used. The filters were flushed with WFI for ≥ 100 L/m2 and flush buffer (50 mM Tris, 50 mM glucose, 10 mM EDTA, pH 7.5) for ≥ 10 L/m2 at 150 LMH before use. Flocculated material was filtered through depth filters at 100 LMH.
Filtration of the product was stopped and switched to the buffer flush when the flocculated material reached the bottom outlet of the bag. We performed a post-product filtration flush at ≥ 10 L/m2 at 100 to 150 LMH to maximize product recovery. The max. pressure observed during filtration was 0.35 and 0.25 bar (for the 50 and 200 L runs respectively. The clarified product was collected into a 200 or 500 L XDUO mixer. For the 200 L run, only 50% of the centrifuged cell concentration material was processed through the lysis and clarification process steps.
Precipitation, flocculation lift, and depth filtration data, buffers, and equipment are shown in Table 6.
Table 6. Precipitation, flocculation lift, and depth filtration procedure for 50 and 200 L fermenter scale
| Step | Procedure | Details | 50 L XDR run | 200 L XDR run |
4 |
Precipitation, flocculation lift, and depth filtration |
Precipitation buffer | 5 M calcium chloride | |
| Flocculation lift buffer | 120 g/L ammonium bicarbonate | |||
| Calcium chloride and flocculation lift buffer addition pump | Watson Marlow 620 series peristaltic pump | Watson Marlow 620 series peristaltic pump | ||
| Flocculate Lift material weight (kg) | 213.0 | 449.2 | ||
| Depth filtration flush buffer | 50 mM Tris, 10 mM EDTA, 50 mM glucose, pH 7.5 | |||
| Depth filtration filters: | 1 × 1 m2 | 2 × 1 m2 | ||
| Depth filtration product filtration flowrate (L/min) | 1.7 | 3.5 | ||
| Depth filtration product filtration weight (kg) | 177.7 | 409.2 | ||
| Depth filtration pump | Watson Marlow 620 series peristaltic pump | Watson Marlow 620 series peristaltic pump | ||
| Depth filtration collection vessel | Xcellerex™ XDUO 200 mixer | Xcellerex™ XDUO 500 mixer | ||
| Depth-filtered product weight (kg) | 196.6 | 437.2 | ||
Analytical methods
pDNA titers were determined for the end of fermentation samples by a DNA miniprep (Qiagen QIAprep Spin miniprep kit) followed by UV-Vis quantitation with a NanoDrop spectrophotometer (Thermo Fisher). Following this application note, we determined neutralized lysate titers using a Sepharose™ High Performance chromatography resin method (PlasmidSelect Xtra screening kit, Cytiva).
We verified the correct plasmid by agarose gel electrophoresis (AGE) +/- pDNA restriction cleavage. Following vendor-recommended protocols, we used the method at the end of fermentation miniprep samples (precast agarose E-gel 1%, Invitrogen and New England Biolabs enzymes).
We determined pDNA SC/OC ratio by AGE and the Invitrogen gels following the vendor-recommended protocol. Imaging was performed with an Amersham™ ImageQuant™ 800 biomolecular imager.
To analyze eluate purity: E. coli residual DNA was determined using qPCR (resDNASEQ quantitative E. coli DNA kits, Thermo Fisher), following the vendor-recommended protocol.
E. coli host cell protein (HCP) was determined via ELISA assay (E. coli HCP ELISA kit, 2G, Cygnus), following the vendor-recommended protocol.
Endotoxin amount was determined via an LAL test, using the Endosafe nexgen-PTS system (Charles River, LAL test cartridges [FDA-licensed], 1.0 to 0.01 EU/mL), following the vendor-recommended protocol.
pDNA SC/OC ratio was determined by capillary electrophoresis (dsDNA 1000 kit, Sciex), following the vendor-recommended protocol.
CY45846