Prolong your ÄKTA system chromatography column lifetime with Protein Prep Syringe Filters
Good practice in liquid chromatography (LC) is to filter samples prior to injection onto the column. Sample filtration prevents unwanted particulates from entering the column. This is important because particulates can reduce column lifetime, increase run time, and distort peak shape. In addition to affecting the quality of your results, particulates might clog the column inlet, causing increased back pressure and premature ending of chromatography runs. These, particulate-related issues can lead to preventable delays and system downtime that reduce lab efficiency. Adjusting protocols to extend usable lifetime can minimize the frequency of column replacement while maintaining data quality throughout.
Although filtration prior to chromatography is common practice, the factors that impact ideal filter selection often are not considered. This lack of consideration can lead to suboptimal results. The ‘right’ filter depends on the method that the sample is being prepared for, the chemical properties of the solvent being used, and the physical and chemical properties of the sample itself.
In this article, we present the results of three analyses demonstrating the suitability of the regenerated cellulose membrane which is contained in the Cytiva Protein Prep syringe filter for protein work. First, we consider the impact of filtration on column cleanliness. Second, we look at proteinaceous sample recovery using regenerated cellulose as a preparation medium. Finally, we present information relating filter quality to the fidelity of results. Regenerated cellulose from Cytiva is batch tested to ensure low levels of extractable compounds that might otherwise interfere with analyses. We also present an overview of solvent chemical compatibility for regenerated cellulose demonstrating its broad compatibility with common aqueous and organic solvents. The analyses presented here provide the basis for our recommendation of the Protein Prep Syringe Filter.
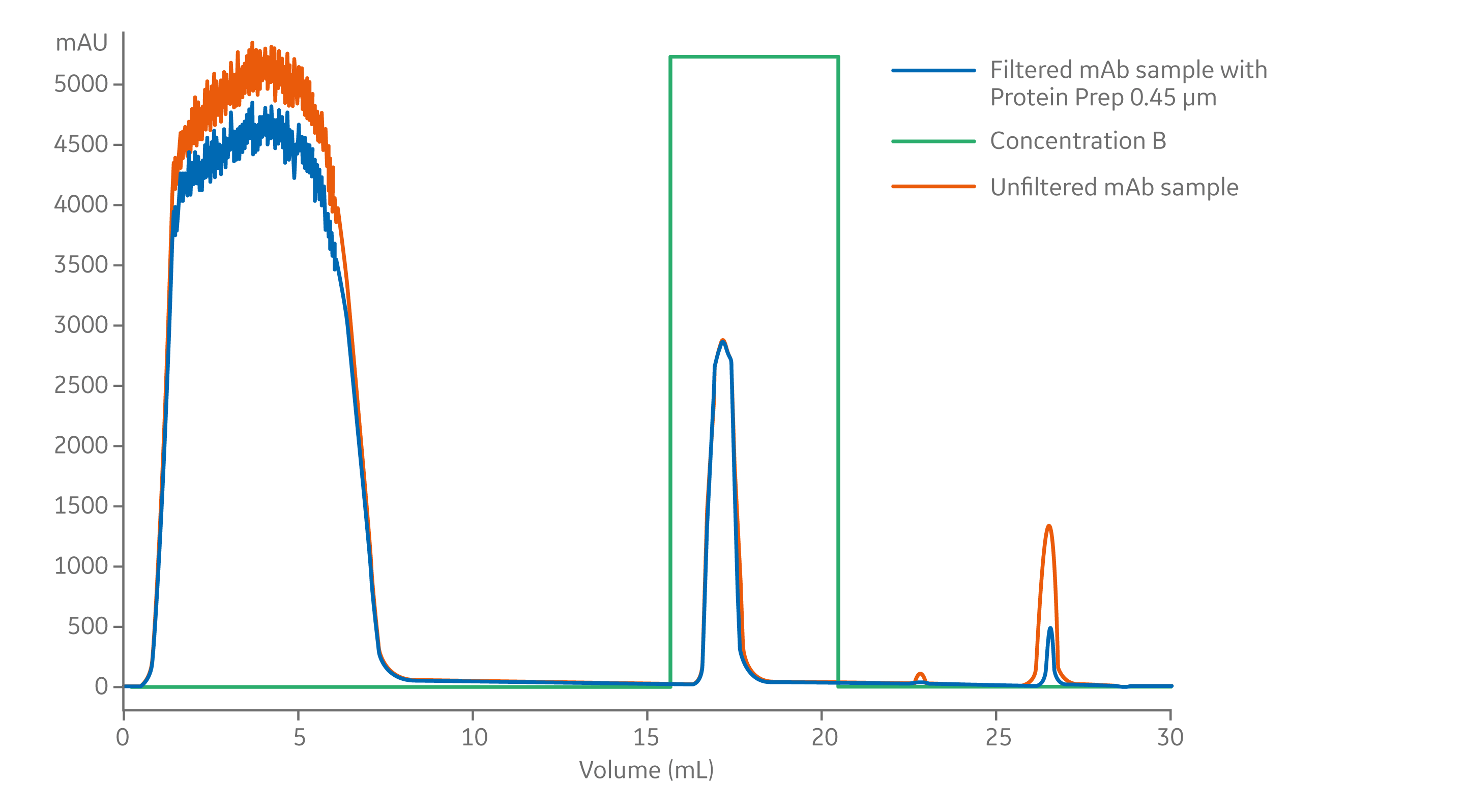
To demonstrate the effectiveness of filtering samples before performing LC, we applied 6 mL cell supernatant containing a human monoclonal antibody onto a HiTrap MabSelect PrismA column, either with or without filtration. The filtered sample was prepared using a 30 mm syringe filter containing a 0.45 µm regenerated cellulose membrane. After the 30-min chromatography run, the column was cleaned with 1 M sodium hydroxide, visible on the chromatogram as a clean-in-place (CIP) peak (Fig 1).
Fig 1. mAb purification on a 1 mL HiTrap MabSelect PrismA column run on ÄKTApure 25 chromatography system. Detection was performed at A280 nm.
Column: HiTrap MabSelect PrismA 1 mL
Sample: 6 mL cell supernatant containing a human monoclonal antibody, either unfiltered or filtered through 0.45 µm, 30 mm regenerated cellulose membrane syringe filter
Binding buffer: 20 mM phosphate, 150 mM NaCl, pH 7.4
Elution buffer: 50 mM phosphate acetate, pH 3.5
Flow rate: 0.5 mL/min
Residence time: 2 min
System: ÄKTApure 25
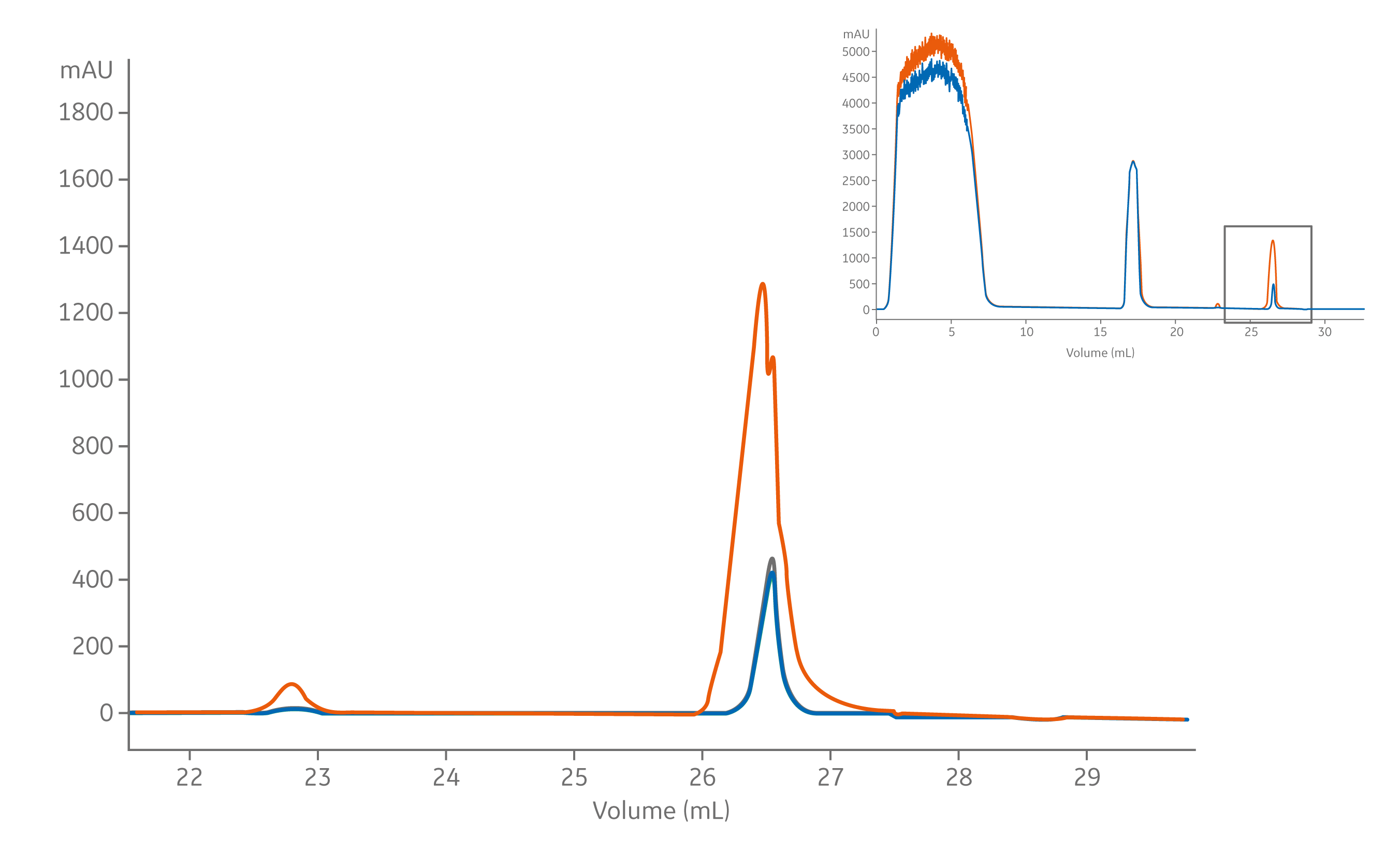
The CIP peaks (~eluted at ~27 ml) was much larger after purification of the unfiltered mAb; indicating that UV-absorbing material remained on the column and was removed with sodium hydroxide treatment. To avoid subjecting columns to extra cleaning cycles, we recommend filtering samples prior to loading on chromatography columns. Filtration to remove particulates can protect the column and maintain the column lifetime (Fig 2).
Fig 2. The clean-in-place (CIP) peak after purification of the unfiltered (purple) and filtered (green and yellow) mAb samples.
The next set of testing concerned sample recovery. The regenerated cellulose membrane demonstrated low levels of nonspecific protein absorption when challenged with two concentrations of a model protein, bovine serum albumin (BSA) dissolved in PBS, pH 7.4 to prepare a 5 mg/mL stock that was further diluted. The results (Table 1) show that well over 95% protein recovery (generally considered to be high) was obtained with regenerated cellulose syringe filters.
Table 1. Percent recovery of BSA samples filtered with regenerated cellulose membranes
| Pore size | Recovery at 1 mg/ mL (%) | SD | Recovery at 0.5 mg/ mL (%) | SD |
| 0.2 µm | 98 | 0.6 | 97 | 0.3 |
| 0.45 µm | 99 | 0.7 | 99 | 0.9 |
| SD= Standard deviation, N=3 | ||||
During an evaluation of sample loss resulting from the filtration process itself (i.e., the amount of sample retained in the device after filtration), the data demonstrated that regenerated cellulose membrane has low sample loss because of the hold-up volume, especially after an air purge. This assessment concluded that standard syringe filtration is appropriate for applications that do not require absolute quantitation of analyte or maximum yield. (Table 2)
Table 2. Hold-up volumes of water in filtration devices containing regenerated cellulose membranes. Results before and after purge
| Pore size | Hold-up volume (µmL) | Final hold up volume (µmL) | ||
| Average | SD | Average | SD | |
| 0.2 µm | 135 | 36 | 9.3 | 0.9 |
| 0.45 µm | 135 | 31 | 9.2 | 1.1 |
| SD= Standard deviation, N= 10 | ||||
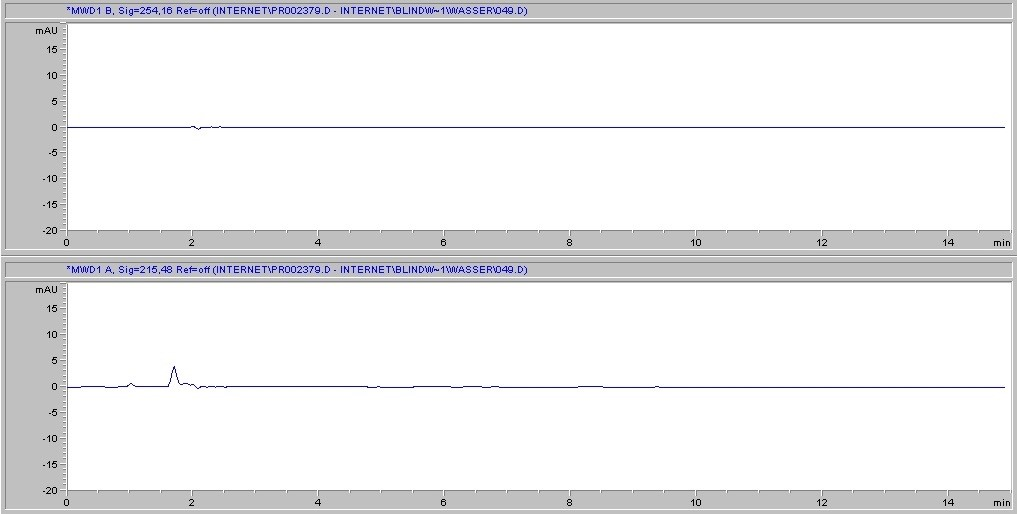
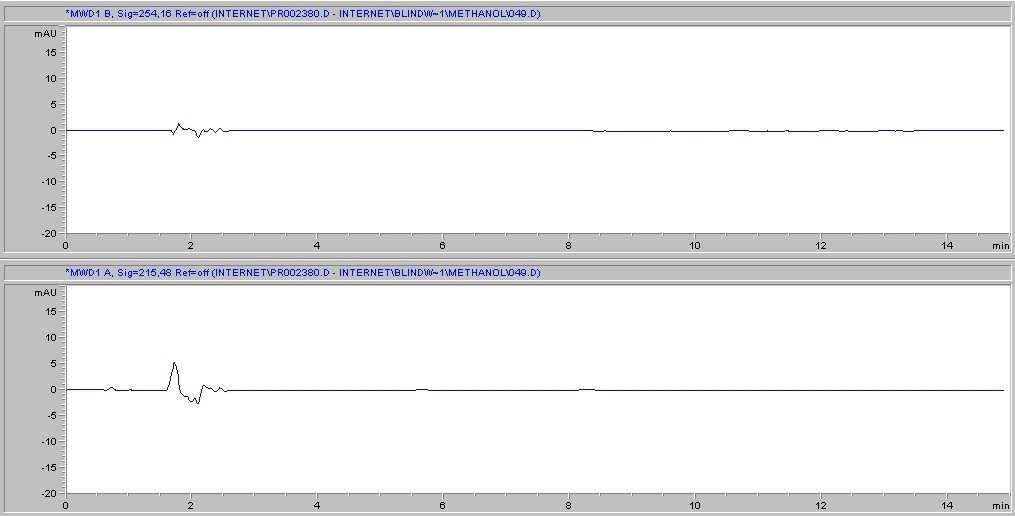
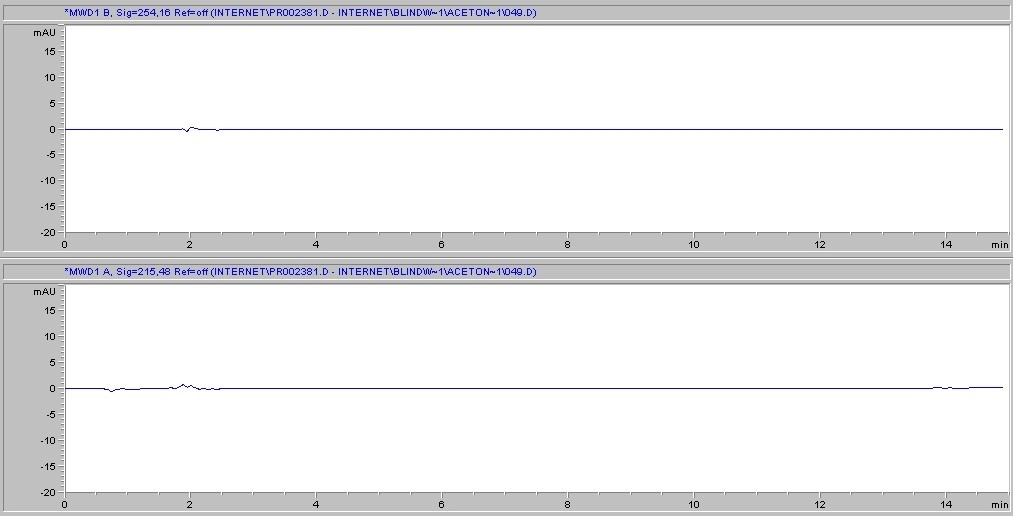
Next, we consider the likelihood of the filter itself to introduce interference into the sample. This can occur if the solvent causes substances to leach out of the membrane. Cytiva's Protein Prep Syringe Filter is certified for the absence of UV-absorbing substances at wavelengths of 210 and 254 nm with water, methanol, and acetonitrile, and has documented batch-to-batch quality and consistency, ensuring reproducible results. (Fig. 3 )Each production lot is tested and certified as such, and certificates of the lot chromatogram can be found at gelifesciences.com/certificates. Moreover, one of the strengths of the regenerated cellulose membrane is its broad chemical compatibility when compared to other common membrane types. Chemical compatibility (Table 3) is meant to assure minimization of both substance leaching and risk of membrane degradation due to interaction with the solvent.
A. Protein prep for ÄKTA systems 30 mm, 0.2 µm – water
B. Protein prep for ÄKTA systems 30 mm, 0.2 µm – methanol
C. Protein prep for ÄKTA systems 30 mm, 0.2 µm – acetonitrile
Fig 3. Representative chromatograms of solvents filtered through regenerated cellulose membrane syringe filters and subjected to HPLC analysis. Top panel in each set of chromatograms shows results at 254 nm; bottom panel shows results at 215 nm.
Although most protein-based work will likely use an aqueous solvent, having a broadly compatible membrane ensures that, in the rare instance when a non-aqueous solvent is used, there will not be a delay introduced into the work due to the need to find a more appropriate syringe filter.
In summary, our analyses determined that regenerated cellulose is well suited for work with proteins, maintaining both column lifetime and data quality. When challenged with two concentrations of a model protein, the membrane demonstrated low levels of nonspecific protein absorption, as well as low hold-up volume in syringe filters. We also showed that it is unlikely for substances to leach out of the membrane, ensuring reproducible results with commonly used solvents.
Therefore, we recommend using Protein Prep Syringe Filters for consistent, accurate, and reproducible filtration prior to liquid chromatography on ÄKTA systems. Protein Prep Syringe Filters are ready-to-use with polypropylene housing and a regenerated cellulose membrane that is broadly compatible with organic and aqueous solvents when compared to other membrane types. They are certified and specifically designed for analytical confidence. Using Protein Prep Syringe Filters can maintain column life and run times. Also, they can protect columns from particulates and avoid the need to replace columns prematurely.
Chemical compatibility of the regenerated cellulose membrane
Table 3. Chemical compatibility of the regenerated cellulose membrane
|
|
|
R = Resistant
LR = Limited Resistance
NR = Not Recommended
* Short-term resistance of housing
The above data is to be used as a guide only.
Testing prior to application is recommended.