-
Protein purification
Access information about chromatography methods, how to set up protocols, and selecting products.
-
Western blotting
Get guidance on efficient transfer, detection, identification and quantification of your target protein.
-
Interaction analysis with Biacore surface plasmon resonance (SPR)
Get insights into key SPR applications, how to get started with SPR interaction analysis, and Biacore products.
Understanding protein production from start to finish
In protein research, going from idea to pure characterized protein takes multiple steps and techniques. When streamlined, this process can help you get results fast, meet publishing deadlines, and make critical decisions on a project.
Most scientists working with exploring proteins and developing specific target biomolecules use several steps including cell culture, sample preparation, purification, and analysis. This requires understanding the principles behind many different tools – and learning these steps takes time that scientists could spend generating data and progressing research. If you’re short on time, setting up a strategy for a complete protein development and production workflow can seem overwhelming.
Scientists at Cytiva Jon Lundqvist, Jinyu Zou, and Anna Moberg have more than two decades of experience in protein research. This article covers their expert guidance, insight, and tips to help you make fast progress in protein research.
From A to Z in the protein production and purification workflow
Over the years, advancing technology has continued to simplify and improve usability across steps in the protein research workflow, enabling individual scientists to cover all or larger parts of it. That’s mostly good news – but it can also mean scientists have less time to learn procedures like cell culture, protein purification, and protein characterization.
To help guide your protein production, Jon, Jinyu, and Anna are combining their expertise to provide insights that can quickly get you up to speed and ready for your next publication or presentation.
Steps in protein research: cell culture, sample prep, purification, and analysis
Before setting up the protocol, set your goals and objectives. To do this, ask yourself questions like:
- What is the purpose of your research? How will you use the purified protein (e.g., to determine structure and function, for therapeutic or diagnostic use, etc.)?
- How much information do you have on the identity and function of the target protein?
- What analysis methods will you use?
- Do you need post-translational modifications (PTM), such as glycosylation, for protein functionality? Will that influence the expression system you choose?
Clear goals help you decide which expression system to use, the purity and yield to aim for, and the amount and quality of sample you need to conduct multiple analyses. Setting goals will also help you choose which analysis methods to use.
When it comes to choosing equipment and consumables, do some extra research. Different proteins have different properties – so reusing protocols and existing equipment might not provide the outcomes you want. Search the literature in your field, and ask around for more insight. Your colleagues can have a wealth of insight depending on the size of your lab. Conferences, meetings, and webinars can be great opportunities to learn from other scientists. Vendors can also provide methods that are easy to replicate or adapt to your application.
Cell culture
Construct design
Once you’ve identified your protein of interest, it’s time to design the DNA template – also known as the construct. The DNA template codes for the protein of interest so that it fits your expression system.
Traditionally, scientists design the DNA template using RNA isolation from relevant host cells (e.g., mouse hepatocytes) followed by cDNA synthesis and PCR amplification of the appropriate gene. The cloning strategy is normally designed within the PCR primers and allows cloning of the PCR insert into a suitable DNA vector to generate a protein expression plasmid.
Nowadays, synthetically made gene sequences are a time- and cost-effective alternative to traditional cloning. Vendors can codon-optimize synthetic genes for the intended expression host cell, increasing the likelihood of obtaining a high expression construct. Most vendors in the synthetic gene space also offer cloning services and can supply ready-to-use protein expression plasmids.
Recombinant proteins (with or without introduced tags in the construct) are expressed in a protein host cell system (e.g., mammalian, prokaryotic, insect, yeast, etc.) through transformation and transfection with the expression plasmid. You need larger amounts of expression plasmids to obtain high levels of protein expression in certain expression systems, especially mammalian cells. To obtain these amounts, you can propagate the expression plasmid in suitable E. coli strains before plasmid purification and before transfecting the plasmid into the expression host cells.
You can use tags to simplify identification, purification, and analysis of expressed proteins. The most common tag is histidine (His-tag) because it’s small and can remain on the protein after purification.
While His-tag is a good place to start, sometimes you’ll want to test one or more alternative tags like GST-tag, Strep-tag II, or MBP-tag. You need to select a tag that’s soluble and that attaches to either the N- or C-terminal side on the target protein – this makes it accessible for detection and binding to the purification resin. If you need to remove the tag after purification, you’ll need to create a cleavage site in the tagged protein vector.
Selecting an expression system
There are several factors to think about when selecting an expression system. If you haven’t already, fill out the goals and objectives for your research we outlined earlier. You can select the expression system based on the purpose of the purified protein, as well as the amount, purity level, and requirement for different variants needed.
You will need to evaluate whether to use a prokaryotic or eukaryotic expression system. E. coli is the go-to-platform for many recombinant proteins – it has well-known genetics and high transformation efficiency, and it’s easy, fast, and inexpensive to cultivate. However, if your protein doesn’t express well in E. coli, then an insect or mammalian system can be more efficient and cost-effective.
In addition to your larger goals, keep in mind some practical considerations:
- Structural complexity: Bacterial proteins and eukaryotic proteins with a simple tertiary structure are good candidates for expression in bacterial systems. Eukaryotic proteins with a more complex tertiary structure are better candidates for expression in mammalian or insect cell systems that can support protein folding and post-translational modifications.
- Post-translational modifications (PTMs): PTMs can be critical for a protein’s folding, function, and localization. These modifications include phosphorylation, glycosylation, prenylation, disulfide bonds, protease cleavage, and more. Mammalian cells have the most sophisticated PTM pathways.
- Solubility: Eukaryotic proteins expressed in bacteria can be biologically inactive and localized to insoluble inclusion bodies because PTM systems are lacking, disulfide bond formation is difficult, and bacterial cells don’t have the same chaperone networks as eukaryotic cells to assist with protein folding. In some cases, you can purify your protein of interest from inclusion bodies using solubilization and refolding steps. If that doesn’t work, then you might want to use expression in a eukaryotic system.
- Cellular localization: A bacterial, insect, or mammalian cell system can be a viable option for recombinant expression of an intracellular protein. The expression of extracellular and/or membrane-bound proteins present additional challenges – in many cases, the PTM and intracellular pathways in eukaryotes make the insect and mammalian systems the go-to option for these proteins.
- Toxicity: Some exogenous proteins’ expression can be toxic to a host cell. To overcome this, you can use special expression vectors and modified E. coli strains. In mammalian cells, you can also consider using inducible systems (e.g., tetracycline-regulated on/off).
Cell culture conditions
To optimize protein expression, you should screen cell culture conditions in 24- to 48-well plates and manage 1 to 2 mL cultivations in parallel.
During the screening, conditions like temperature and additives vary. If the expression levels are lower than you expect, or if the target protein is not expressed, you should go back and look at factors like the tag system and cultivation conditions. Optimize them until you find the clone and expression levels that meet the goals you set.
Once you’ve chosen an expression system and the conditions to grow the host cell, you can scale up to larger cell culture volumes. Scientists commonly use cell culture volumes from 0.1 to 20 L in research scale, and use shaker flasks when working with E. coli.
For higher expression levels and mammalian cells, you can use a fermenter to better control conditions like temperature, oxygen, glucose levels, and pH. Exact conditions vary from cell type to cell type, but all cultures have a vessel and medium that support cell growth by supplying hormones, growth factors, and nutrients. To prolong mammalian cell viability and growth, use specific medium feeds in conjunction with chemically defined media, or supplement classic formulations with serum.
The physicochemical environment (pH, osmolality, etc.) is also critical to successful cell culture – you can maintain it by using process liquids and buffers.
Sample prep
Throughout sample preparation, protein purification, and analysis, you need to avoid protein damage and ensure that proteins maintain activity.
Some proteins are more sensitive than others. Understanding your protein will help you determine which conditions to use and which to avoid. Some proteins, like secreted proteins, have lower expression levels – when expression levels are low, it becomes even more important to make sure you’re maintaining high yield in each step.
Sample preparation depends on protein type. Membrane proteins, protein complexes, and secreted proteins will have different sample preparation protocols. For intracellular proteins, scientists lyse the cells to release the target – you can do this through sonication, homogenization, freezing and thawing, or chemical agents.
For extracellular proteins obtained with mammalian or insect cells, you can directly apply cell harvest. Scientists often perform cell harvest using centrifugation. In most applications, scientists transfer the supernatant to protein purification containing the target protein, or analyze it directly to perform identification and determine activity.
If needed, you can use filtration to remove particles after centrifugation, or before chromatography to protect your columns. Desalting at laboratory scale is a well-proven, simple, and fast method that removes low molecular weight impurities and transfers the sample into required buffer in a single step.
You can use desalting or buffer exchange prior to a chromatography step or between purification steps for sample conditioning.
Protein purification
In protein research, scientists often use two purification steps — affinity chromatography and size exclusion chromatography. If you need high purity, add an additional intermediate step of ion exchange or hydrophobic interaction chromatography. However, try to use as few steps as possible — adding steps decreases overall protein yield.
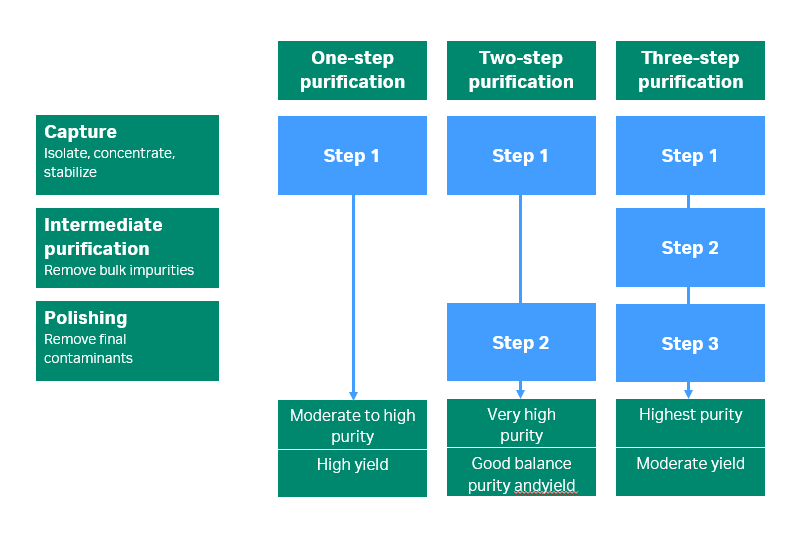
Tagged recombinant proteins are usually straightforward to purify. Use an affinity resin that’s suitable for the tag system you’re using, and use the protein’s natural conditions to avoid precipitation and degradation.
You might need to remove the tag after purification. After tag cleavage, you need to separate tag-free target protein from the tag and tag-containing target protein. You can also use on-column cleavage – add protease to the column with the target protein bound to enable one-step cleavage and removal.
Scientists often use affinity chromatography for antibody purification. Affinity chromatography uses the high affinity and specificity of protein A and protein G for the Fc-region of IgG from many species. Protein L, which binds to kappa light chains, is another ligand you can use to purify antibody fragments, IgG, and other antibodies from a wide range of eukaryotic species. For higher purity, scientists often use SEC as a second step.
You’ll need more effort when developing the protocols for nontagged proteins. Experiment planning helps you decide which conditions to test, as well as what to optimize to achieve your desired purity and yield.
Scientists commonly use gravity columns in affinity steps and occasionally use peristatic pumps in other chromatography steps. A protein purification system, often in combination with a prepacked column, delivers significantly more control, enabling you to obtain more detailed and consistent information on your target protein and any impurities. It also provides better column protection.
Protein analysis – identification and characterization
Proteins are diverse in size, structure, and biochemical properties. Scientists use these differences to characterize isolated proteins. You can use multiple techniques to confirm identity and binding activity during protein expression, production, in between steps, and within the purified protein.
A lot of techniques and methods are common in analysis and characterization. UV, SDS-PAGE, and SEC are popular choices for identifying which fractions contain the target protein. Assays for specific proteins like enzymatic assays are available and require a substrate specific to the target protein.
After purification, you can determine structures, binding activities, stability, and more. That’s often done using mass spectrometry in combination with high-performance liquid chromatography (HPLC) columns, amino acid sequencing, X-ray crystallography, cryo electron microscopy (cryo-EM), surface plasmon resonance (SPR), Western blotting, circular dichroism, and nuclear magnetic resonance (NMR) spectroscopy.
Western blotting
Western blotting is one of the most established and popular techniques for quantifying and identifying proteins. The method builds an antibody:protein complex – scientists bind the antibodies to membrane-immobilized proteins, and detect the bound antibody with a detection method like chemiluminescence or fluorescence. A sample can be a complex protein mixture like a cell or tissue extract, but it can also be a sample of purified proteins, like a fraction from a purification run.
The Western blot workflow has several steps before analysis:

Scientists apply gel electrophoresis to the sample for protein separation before immobilizing the proteins on a membrane, (e.g., Nitrocellulose or PVDF) following electrotransfer from the gel.
Scientists then incubate the membrane with a primary antibody that specifically binds to the protein of interest, wash to remove unbound antibodies, and conjugate a secondary antibody to an enzyme, a fluorophore, or an isotope for detection. The detected signal from the protein:antibody:antibody complex is proportional to the amount of protein on the membrane.
Traditional total protein staining methods often limit the dynamic range, typically by one or two orders of magnitude for silver and Coomassie staining. In contrast, fluorescence and chemiluminescence detection has a much broader detection window and is quick, easy to use, and highly sensitive.
You can use a CCD-based imager to visualize the blot in both chemiluminescence and fluorescence ranges with high sensitivity and resolution.
Determining mechanism of action using SPR
Understanding the nature of molecular interactions is critical in all areas of the life sciences. Label-free interaction analysis generates data-rich content that can help enhance our understanding of interactions between almost any type of biologically relevant molecules.
- Do the potential interactants bind to each other?
- How specific is the interaction?
- How strong is the binding (i.e., affinity)?
- How fast is the binding?
- What are the effects of temperature?
- How much interactant is there in the sample?
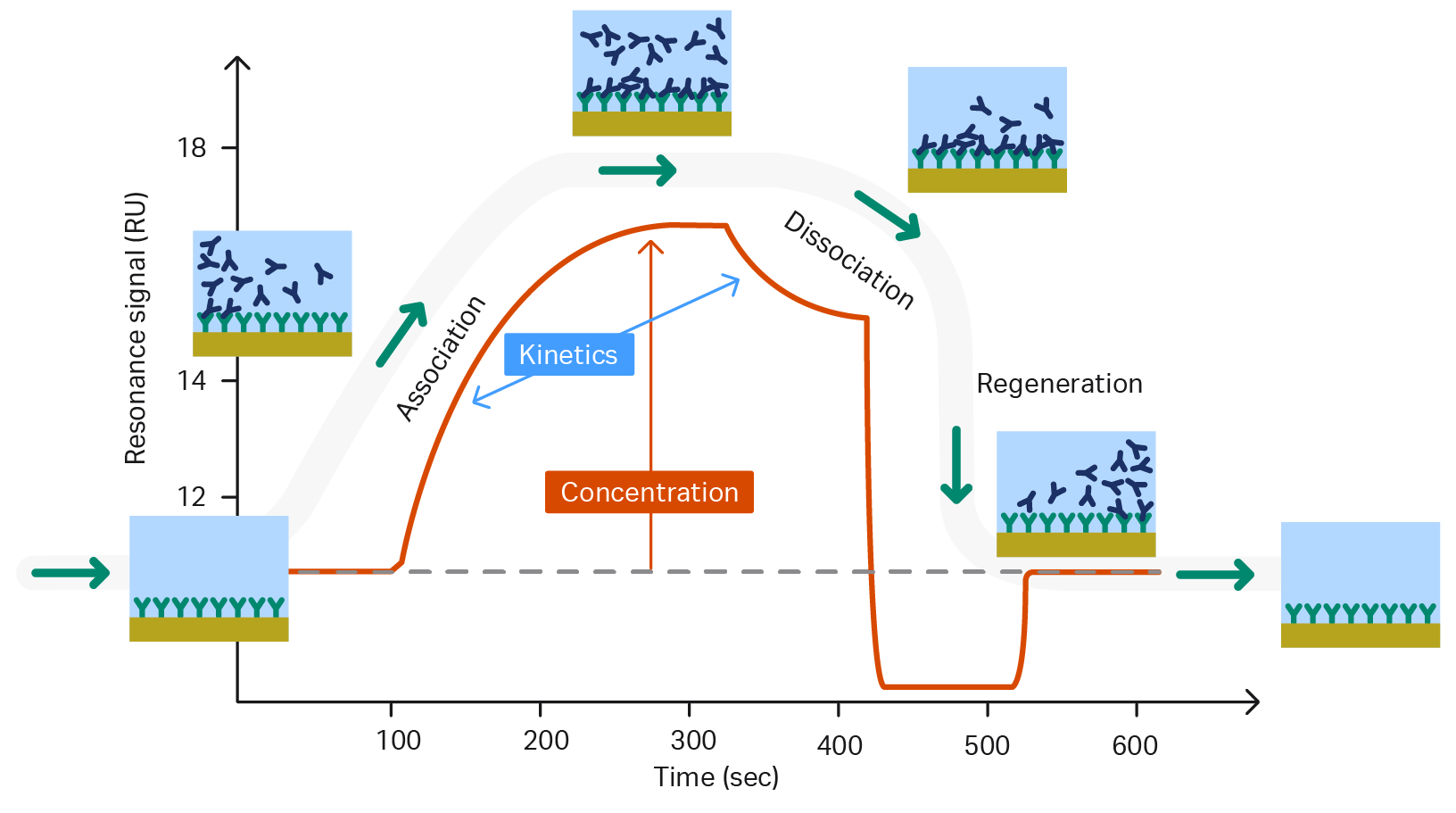
Biological processes are real-time events that are driven and regulated through dynamic interactions between key molecules. End-point techniques like ELISA can only offer a snapshot, and provide basic information like overall binding strength. Affinity depends on the ratio of on- and off-rates, so equal affinity interactions can have very different kinetic properties. Biacore systems enable real-time analysis that can provide the data needed to discriminate these crucial differences.
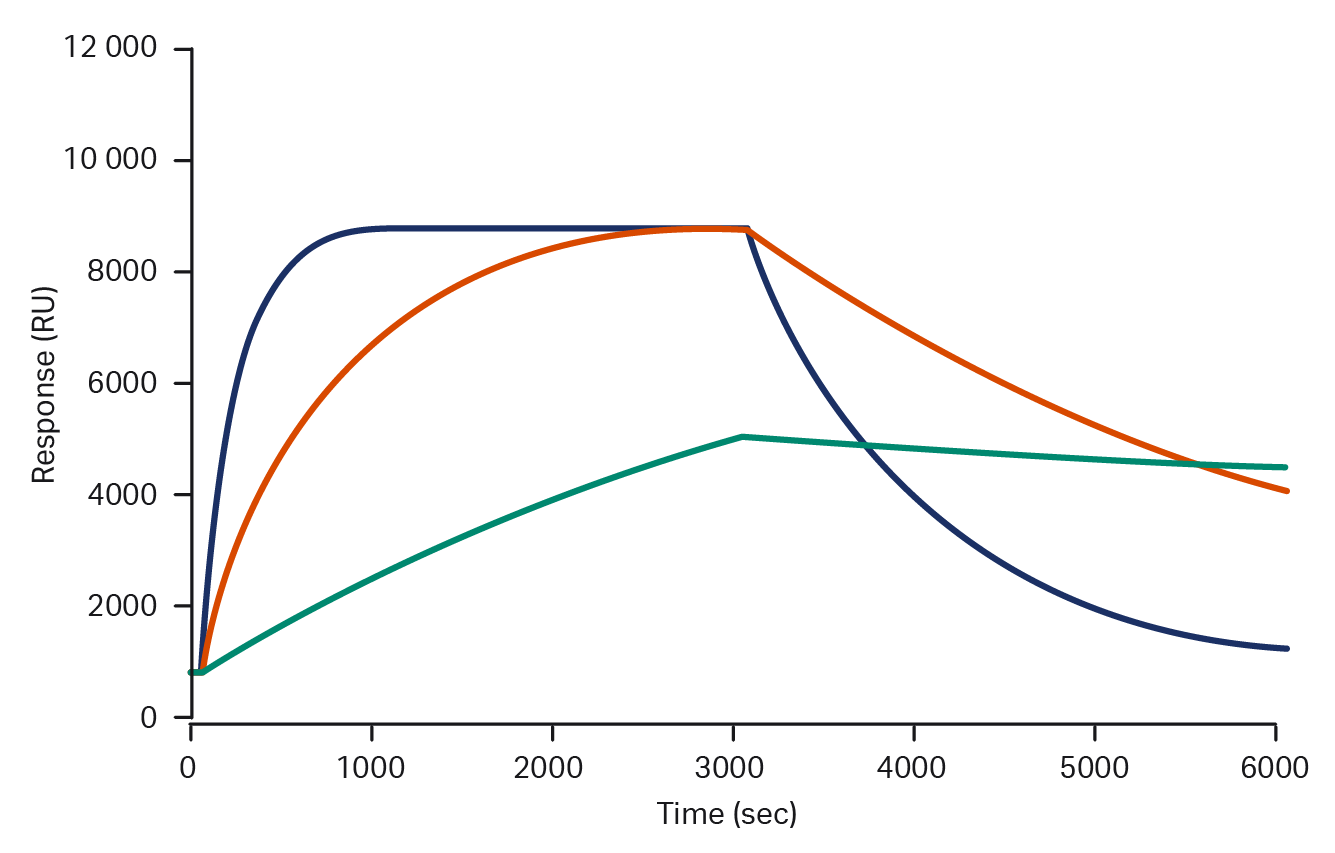
Sensorgrams showing three molecules with identical affinities, but kinetic profiles differing by several orders of magnitude. These differences are not visible with end-point analyses.
Biacore sensor chips support analysis of a wide range of interactions. You can use kits and ready-made consumables to save time while ensuring consistent capture of molecules e.g. antibodies and common tags.
We recommend using predefined Biacore run methods with application-relevant settings. Combine them with evaluation software for a good data overview that shortens time to results.
Guidance and tips
You might want a one-size-fits-all approach to your protocols, but this can cost you time in the long run. You need to adjust protocols to fit specific proteins, and add, change, or remove steps for specific targets. One-protocol-fits-all thinking can lead to low expression, poor yield, and time-consuming troubleshooting.
We’re often eager to get started and don’t spend enough time reading the literature or planning. While fast at first, suboptimal protocols will eventually delay our results. Defined goals, clear objectives, and thorough planning will help you get better results faster.
Access more knowledge about the different steps in protein production by clicking on the different categories below or sign up for our newsletters.