Introduction to Cytiva™ Protein Select™ technology and self-cleaving tag
Purifying a recombinant protein with its native sequence in a single chromatography step is challenging if the target protein does not have a natural affinity binding partner. Even then, traditional affinity elution conditions are often harsh, and can lead to a loss of activity and recovery of the target protein.
The Cytiva™ Protein Select™ technology is an affinity chromatography system that uses a self-cleaving tag. This is beneficial for industry applications where high purity of target proteins derived from complex samples are needed without having residual, potentially interfering, affinity tags remaining.
The technology enables the production of purified, tag free proteins in a single step for predictable results (see Fig 1).
Fig 1. Overview of Cytiva™ Protein Select™ affinity purification.
An added benefit is that no proteases are needed in this process. Proteases are often inefficient and non-specific, resulting in the need for further purification steps after cleavage.
The objective of this article is to explore the hold step (shown in Fig 1) and the main factors that impact cleavage efficiency when using Cytiva™ Protein Select™ technology.
Description of the cleavage mechanism
The minimum Cytiva™ Protein Select™ tag sequence is 36 amino acids; the sequence may vary slightly depending on the expression system used. Only the C-terminal (second half) of the Cytiva™ Protein Select™ tag is directly involved in the cleavage reaction. The first requirement for cleavage is the initial binding of the tag to the ligand with high affinity. Upon binding, the tag and ligand fold and create a catalytic core that induces cleavage of the target protein at the last amino acid of the tag (Asn). Bound target proteins are released by this automatic cleavage when ligand-tag complex has been formed, while the cleaved tag remains on the resin, as shown in Figure 2.
The reaction is spontaneous and does not require external stimulus, harsh conditions, or addition of excipients. Folding is the rate-limiting step since binding is very fast and only limited by diffusion.
Fig 2. Self-cleaving complex formed between ligand and tag.
Cleavage can only occur once a complex has been formed between the Cytiva™ Protein Select™ ligand and tag. This predominantly occurs during the hold-step during purification.
Several factors affect the cleavage efficiency and subsequently, the final cleavage yield. These are:
- Amino acid residues at the cleavage site
- Temperature of the cleavage reaction
- Duration of the cleavage reaction
- pH of the cleavage buffer
- Size and shape of the target protein
The N-terminal sequence of the target protein strongly affects the cleavage efficiency
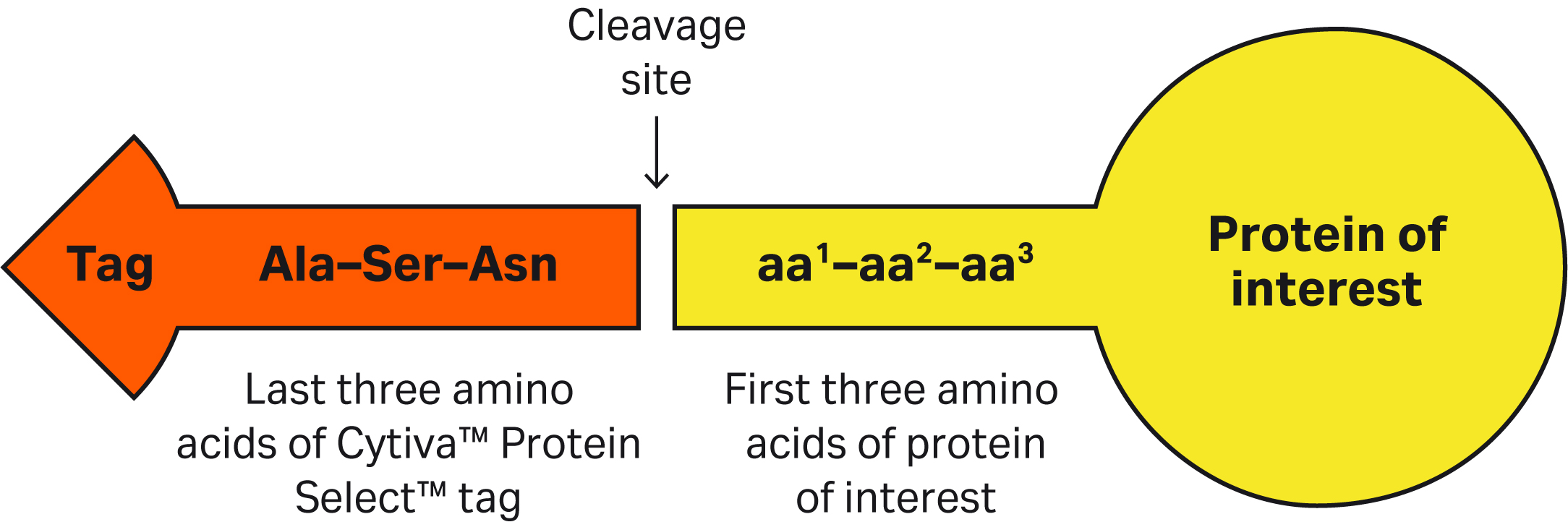
Amino acid residues of the target protein (Fig 3) will affect the active-site formation and spatial arrangements of catalytic amino acids in the ligand-tag complex. The first three amino acids of the target protein heavily influence the cleavage kinetics and can result in slower or faster cleavage. It is not recommended to have Proline in the first or second position of the target protein as it causes an extremely slow cleavage reaction. Other amino acids may enhance or slow down the cleavage kinetics depending on the position at the N-terminus. The combination of amino acid residues in positions 1 and 2, that are closer to the active-site catalytic residues, display an additive effect on cleavage kinetics.
Fig 3. Schematic of the cleavage site between the Cytiva™ Protein Select™ tag and the target protein.
The impact of the two first amino acids on the cleavage efficiency was tested using synthetic peptides having the Cytiva™ Protein Select™ tag sequence followed by the variable aa1 and aa2 and a short linker sequence VGGSGK, (wherein the C-terminal K was biotinylated). The choice of six amino acids is depicted in bold text in Table 1, representing the overall characteristics of amino acids in terms of physical properties e.g., size, polarity, charge. The six chosen amino acids were varied in both positions to generate 36 peptides covering all combinations. The synthetic, biotinylated peptides with varying amino acid combinations in the aa1 and aa2 positions were each immobilized to Streptavidin sensor chips for cleavage kinetic studies by injecting free Cytiva™ Protein Select™ ligand to the sensor chips. The study was performed at constant temperature and pH (25°C and pH 7.4), using the HBS-EP+ running buffer.
Table 1. Amino acids chosen for the variable positions, aa1 and aa2 in synthetic peptides are shown in bold text
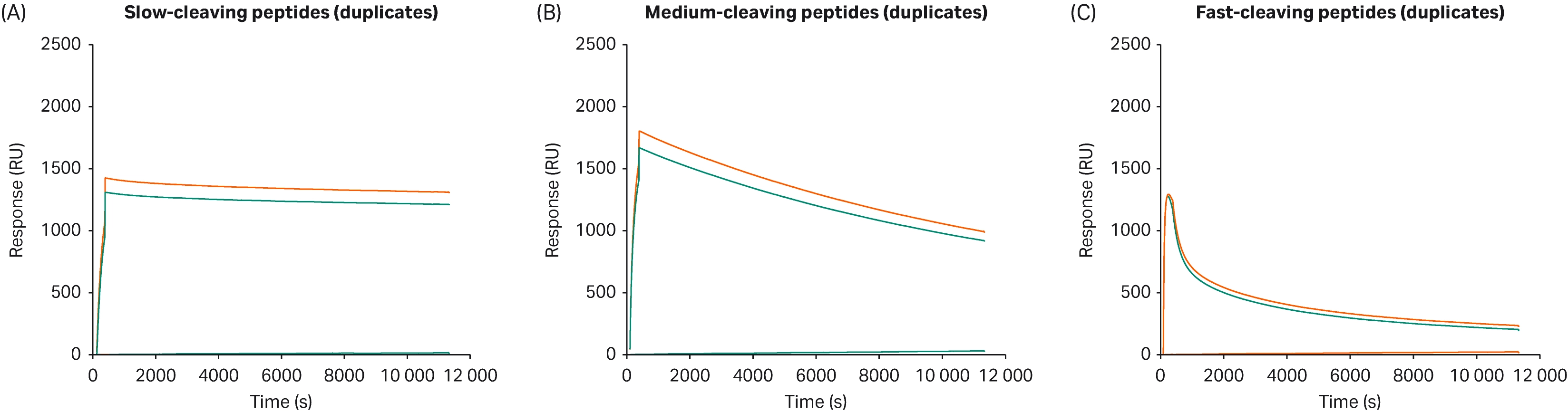
After immobilization of each of the 36 synthetic peptides to the Streptavidin chips, the free Cytiva™ Protein Select™ ligand was injected to the sensor chips for one min, resulting in binding of ligand and immobilized tagged peptides, followed by folding into active complex. The relative binding levels were measured from the increase in response units in the sensorgrams. After ligand injection, the running buffer flowed over the Streptavidin chips for an extended time (30 min up to 3 h). The decline in response units during the wash with running buffer is proportional to the mass lost due to cleavage and can be used to measure cleavage kinetics in real-time. A control experiment with a peptide containing the inactive N36A point mutation of the tag showed no significant decline in response units up to 3 h. The sensorgrams could then be compared to each other and categorized as slow, medium or fast cleaving peptides, as shown in Figure 4.
Fig 4. Sensorgrams showing relative binding levels after one min injection of ligand and then, A) slow, B) medium and C) fast cleaving peptides.
The results of real-time cleavage-kinetics study are summarized in Table 2. The overall conclusion is that larger hydrophobic amino acids in the aa2 position leads to faster cleavage kinetics, whereas a negatively charged amino acid generally leads to slower kinetics irrespective of position. The first position of a target protein (aa1) seems more tolerant to amino acids of various types when compared with the second position (aa2), in terms of cleavage kinetics and efficiency.
Table 2. Analysis data from a real-time cleavage-kinetics study using Biacore™ SPR
Real-time cleavage-kinetics analysis was undertaken using Biacore™ SPR. Identical Cytiva™ Protein Select™ tagged synthetic peptides were used, except for the amino acids at the first two positions after the cleavage site, aa1 and aa2. Identical conditions were used for other parameters, such as temperature, pH, and sample loading amounts.
Cleavage is also dependent on the target protein structure and not only the amino acid residues at the cleavage site. Therefore, the impact of various amino acid residues at the cleavage site according to Table 2, serve as a guideline. In most cases, incubation of a column during the hold-step at room temperature (19°C to 23°C), overnight (16h to 20 h), results in more than 50% cleavage of the Cytiva™ Protein Select™ tagged target protein.
Pulsed elution chromatography to evaluate cleavage efficiency
In this study, we examined the dependency of N-terminal amino acids on cleavage efficiency in test proteins using pulsed elution chromatography. Based on the data obtained (Table 2), we selected the following N-terminal amino acid combinations with varying cleavage efficiency for further study: AF (Ala-Phe), SS (Ser-Ser), SE (Ser-Glu) and RF (Arg-Phe). According to the data in Table 2, AF and RF are expected to exhibit faster cleavage, while SE is expected to exhibit slower cleavage. Human Interleukin-1β (IL-1β) was used as a test target protein and genes with the selected combinations were synthesized with the Cytiva™ Protein Select™ tag for further testing.
The four test proteins were expressed in E.coli and purified via an additional C-terminal TwinStrep tag to obtain pure protein for chromatography experiments.
The purified test-proteins were applied to 1 mL HiTrap™ Protein Select™ columns for cleavage efficiency studies on an ÄKTA pure™ chromatography system. The test was performed at room temperature (22°C), using PBS-buffer (pH 7.4) for all chromatographic steps, except for regeneration/CIP. After sample loading and column washing, the flow was stopped to allow for cleavage, followed by elution of tag-less protein. This was repeated six times to gather data for the time-dependent cumulative yield on-column. The time-points and an example chromatogram showing the elution steps (peaks B-G) are shown in Figure 5.
| Peak | Step | CV |
| Equilibration | 10 | |
| Sample application/wash | 6 | |
| B | Elution 0-15 min | 2 |
| C | Elution 15-30 min | 2 |
| D | Elution 30-60 min | 2 |
| E | Elution 1-2 hour | 2 |
| F | Elution 2-3 hour | 2 |
| G | Elution overnight 12 hours | 5 |
| Regeneration water | 5 | |
| Regeneration urea/NaOH | 10 | |
| Regeneration water | 5 | |
| Equilibration | 10 |
Fig 5. Time-intervals in chromatography method designed for kinetics study and an example chromatogram showing the pulsed elution peaks.
Peak fractions were analyzed with size exclusion chromatography and the amount of cleaved and eluted tag-less protein was plotted in a graph showing the cumulative yield over time, as shown in Figure 6.
Fig 6. The cumulative amount of eluted IL-1β compared with the loaded amount, expressed in %. Contribution from each peak (B-G), indicated in Figure 5. Test protein was IL-1β variants with different N-terminal substitutions in positions aa1 and aa2.
The data from the SPR analysis and the data obtained from the chromatography experiments were compared and show that cleavage kinetics are correlated/related to the amino acids in aa1 and aa2. The time taken for sample application and subsequent column wash was kept to a minimum in these chromatography runs as loading time potentially has a large effect on the cumulative yield, especially for fast cleaving variants.
Previous tests show that cleavage efficiency depends on the N-terminal amino acids of a target protein. Large effects can be seen by small nonpolar or negatively charged amino acids, that are predicted to give slower cleavage rates (see Table 2). An exception is Cys, which results in faster cleavage when found in position aa1 (data not shown). The other large effect comes from bulky hydrophobic amino acids that generally give faster cleaving rates.
Based on our experience, faster-cleaving proteins typically achieve sufficient cleavage employing a 6 h hold time. However, careful attention must be given to sample applications and wash time to minimize sample loss. For medium-cleaving proteins, a hold time of 6 h to 8 h is recommended to achieve an acceptable yield, although an overnight hold may further improve the yield. For slower-cleaving proteins, overnight hold time is necessary, but even with this extended duration, the yield may still be insufficient.
The temperature and duration of cleavage has a direct effect on the cleavage efficiency
In a series of experiments using the test-protein IL-1β, the cleavage yields at room temperature (22°C), were compared with the yields obtained in a cold room at 4°C.
The main chain of IL-1β, 117-269, starts with the sequence APV. Since proline in the aa2 position may prevent cleavage, a point mutation was introduced along with the insertion of the Cytiva™ Protein Select™ tag to generate the mutant designated: Protein Select tagged IL-1β (P2F). According to Table 2, the cleavage-kinetics are expected to be faster.
The test protein was expressed in E.coli and purified via conventional chromatography using a multimodal chromatography step followed by a cation exchange chromatography step to generate pure, intact tagged test-protein. LC-MS analysis of the intact tagged test-protein matched the theoretical molecular weight minus initial amino acid Met of the Cytiva™ Protein Select™ tag, (24978 Da).
The purified test-protein was spiked into conditioned media from CHO-cell cultivations at a final concentration of 1.0 mg/mL. The spiked samples were then applied to 1 mL HiTrap™ Protein Select™ columns to study the cleavage efficiency using ÄKTA pure™ chromatography systems placed in either a cold room or at room temperature. The test was performed using PBS-buffer pH 7.4 for all chromatographic steps, except for regeneration/CIP. After sample loading and column washing, the flow was stopped to allow for cleavage during the hold-step, followed by elution of tag-less protein.
The theoretical molecular weight of intact tagged test-protein is 24978 Da. After cleavage, the theoretical molecular weight of IL-1β (P2F) is 17640 Da. The concentration of tagged test-protein spiked in CHO-feed is 1.0 mg/mL, this corresponds to 0.706 mg/mL IL-1β (P2F), (17640/24978). Calculation of yield (%) will therefore only include recovery of the target protein, IL-1β, since this is what is expected after elution.
Chromatography results
In the comparison of cleavage efficiency at cold room temperature versus room temperature, (measured as yield of eluted tag-less IL-1β (P2F)), the same amount of spiked sample was applied to columns run at different temperatures. The yield (%) of cleaved IL-1β from this experiment (see chromatograms in Fig 7) was 63% at 22°C and 49% at 4°C after using 4 h hold-steps.
The cold room experiment was repeated once using a 20-hour hold-step for the comparison of cleavage efficiency due to different incubation times. The yield was 63% at 4°C using a 20 h hold-step when using the same amount of spiked sample (see chromatogram in Fig 7).
Thus, for this test-protein, the yield (%) from a 4 h hold-step at 22°C was the same as the yield (%) from a 20 h hold-step at 4°C. The cleavage kinetics for this protein follow a double exponential function as shown in Figure 4C, which means that most of the cleavage occurs in the beginning of the hold-step.
Fig 7. Overlay of three chromatograms from the purification of IL-1β (P2F) on HiTrap™ Protein Select™ columns. Purification was undertaken at either room temperature (orange), or in a cold room (blue and green). Lengths of the hold-steps were either 4 h (orange and blue) or 20 h (green). All other conditions and sample loads were the same. The results show that the longer hold-step is required at the lower temperature to reach the same yield obtained at room temperature.
The impact of pH on cleavage efficiency
pH plays a role in cleavage efficiency, although the level of impact is target protein dependent. We tested the pH impact on cleavage efficiency by using IL-1β (P2F) as a test-protein that has a theoretical pH of 7. There was a difference in cleavage efficiency when comparing different buffers in Biacore SPR analysis (see Fig 9). The difference between cleavage at pH 6.0 in this case and 7.4 was larger than the corresponding difference between pH 7.4 and pH 8.8 at a temperature of 25°C and cleavage time of 1 h.
Fig 8. Data from a real-time cleavage-kinetics study using Biacore SPR analysis. Cytiva™ Protein Select™ tagged IL-1β (P2F) was used as test-protein to study cleavage during 1 h at 25°C using different running buffers with different pH.
Summary and recommendations
The most important factors that affect cleavage efficiency and the final yield of a target protein have been discussed in this article. When designing a purification scheme to optimize yield of the target protein there are several things that should be considered.
Sample application time and column washing should be kept shorter to enable faster cleaving of the target protein. Also, the temperature should be kept lower, if possible, during those initial steps to slow the cleavage reaction and improve yield.
For a slower cleaving target protein, a longer hold-step and higher temperature will increase the cleavage efficiency. If the yield is still poor, consider altering the N-terminus of the target protein. This could be done by a point mutation, by substituting amino acids with more favorable cleavage kinetics into the aa1 and aa2 position of your target protein (see Table 2). In this work, the test-proteins having SS or AF at the cleavage site seem to give balanced performance for chromatographic purposes.
Furthermore, the temperature has a direct effect on cleavage efficiency. The comparison of cleavage efficiency at different temperatures and hold-step durations showed that higher temperatures (22°C) resulted in higher yields compared to lower temperatures (4°C). At a low temperature, a longer hold-step should be applied to compensate for the temperature difference.
Finally, the pH could be an important factor, but we should also consider the stability of the target protein or buffer conditions that are suitable or even required for the next step in a process.
RELATED RESOURCES
- FAQ for Cytiva™ Protein Select™ technology
- Data file for Cytiva™ Protein Select™ resin
- Tag sequence for Cytiva™ Protein Select™ tag
- Product page for Cytiva™ Protein Select™ resin and HiTrap™ Protein Select™ column
- Instructions for use for Cytiva™ Protein Select™ resin
- Instructions for use for HiTrap™ Protein Select™ columns
- Integrating Cytiva™ Protein Select™ tag into an expression vector
- AxiChrom™ column packing with Cytiva™ Protein Select™ resin
- Design flexibility of protein construct with Cytiva™ Protein Select™ tag