Magnetic beads for next-generation sequencing (NGS)
Mag beads have many uses within NGS workflows. In addition to nucleic acid isolation and purification, magnetic beads are also used for size selection in NGS library preparation and library normalization. Straptavidin-tagged mag beads are also used for target enrichment and hybrid capture.
DNA and RNA extraction using the chaotropic salt chemistry
Silica resins or silica-coated magnetic beads use chaotropic salts to disrupt hydrogen bonds and bind nucleic acids, enabling contaminants to be washed away. The chaotropic agents help the DNA dehydration and bridge the bead surface and the nucleic acid with their divalent cations. This allows the negative charges of both the bead surface and nucleic acid to be overcome and it is a reversible mechanism.
- Guanidinium thiocyanate, often added to lysis buffer, is a strong denaturant that inhibits RNase and DNase activities.
- Dithiothreitol reduces protein activity, and glycogen helps precipitate nucleic acid.
- Nucleic acid binding to beads is enhanced under high salt concentration, increased incubation time and increased bead volume.
- Chaotropic agents help DNA dehydration and binding to bead surface.
The functional coatings on the beads work via electrostatic interactions or salt- or pH-mediated charge switchable attractions (Fig 1). Specific nucleic acid subsets can be isolated by attaching target oligonucleotides to the bead surface.
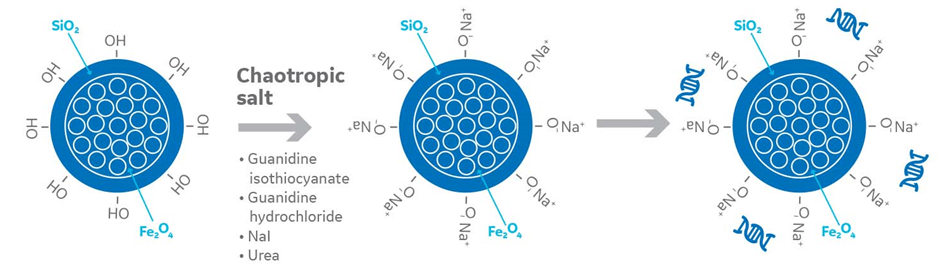
Fig 1. Chaotropic agents in nucleic acid extraction with magnetic beads
Following binding, beads are washed with different concentrations of an ethanol-based buffer to remove contaminants. Stringency of buffer and washes can be increased depending on the level of contamination.
The nucleic acid is extracted in a suitable elution buffer or water by incubation at a high temperature (65˚C for 5 minutes or 80˚C in certain cases) for better recovery (1). In high-throughput automation methods, the elution buffer can be heated prior to adding to the beads to increase elution efficiency.
PCR-amplified DNA products, plasmid DNA, RNA, and viral or bacterial nucleic acids can be purified for downstream applications by bead-based methods. Purity will be affected by the conditions mentioned above. Refer to the manufacturer’s recommendations for specific protocols.
Magnetic beads for next-generation sequencing (NGS) library preparation
NGS library preparation encompasses
- DNA fragmentation
- Addition of adapter sequences
- Size selection
- Library QC
Size selection is a critical step that determines downstream success and can be achieved by magnetic bead-based selection. Typically, large fragments (> 400 bp) that interfere with clustering are removed first, followed by removal of all fragments below 150-200bp. The fragment size of interest is then bound to beads, washed and eluted. Size selection can be influenced by DNA fragmentation; if the sonication protocol used is inappropriate, selection can be inefficient.
By using different volume ratios of bead suspension to sample, different fragment sizes can be isolated, giving users flexibility for bespoke protocols. This can vary among beads, so refer to the manufacturer’s protocol to achieve the best results. Incubate the bead-sample mix for at least five minutes to allow effective binding.
Perform washes while the beads remain in the magnetic field. Residual alcohol from wash buffer can interfere with sequencing, so air-dry samples at the end of the wash protocol to prevent carry-forward. In RNA-seq protocols, magnetic beads are also used to remove particular RNA types, like rRNA, or enrich specific types like mRNA, depending on the particular experiment.
Library normalization with magnetic beads
The idea behind magnetic bead-based normalization is that a given volume of beads can bind a consistent quantity of nucleic acid molecules. That is, if there are enough molecules in each library to saturate the beads, an essentially equimolar quantity of library fragments will bind and be retained from each sample (Fig 2). All unbound molecules are then washed away so that each library is represented by just the bead-bound molecules. The normalized sample is then eluted from the beads for inclusion in the library.

Fig 2. A given volume of beads can bind a consistent quantity of nucleic acid molecules in a four-step process from sample to elution. First, the same volume of magnetic particles is added to each sample and binds to target molecule. Then, a magnetic field is applied to capture the beads with the bound target, and the remainder of the sample is washed away. Finally, the normalized sample is released from the magnetic particles for inclusion in the library.
There are several coating options available to suit any given application: carboxyl- and silica-coated magnetic beads for generic, non-specific binding based on buffer conditions; oligo(dT)-coated beads for binding mRNA; and streptavidin-coated beads for binding biotinylated samples.
This approach is reasonably straightforward, and studies in recent years have indicated that bead-based normalization produces more consistent read depth than several existing quantitation-based methods. Illumina has exploited this approach for normalization, modifying its transposon-based “tagmentation” system for NGS library prep to use magnetic beads.
The bead-based approach, however, can be wasteful: the number of molecules in each library needs to equal or exceed the binding capacity of the beads, with the excess discarded. If your sample is precious or in short supply, it might be worth taking the extra time for qPCR-based quantitation.
For more information about the importance of DNA normalization and the range of methods, see our blog: DNA library normalization for NGS: why and how.
Target enrichment and hybrid capture using streptavidin beads
Streptavidin-tagged magnetic beads are being increasingly used in exome and targeted sequencing due to its strong affinity for biotin (it is known to be the strongest noncovalent biological interaction). Sheared genomic DNA is captured with a pool of target-specific biotinylated probes against the whole exome, target genomic regions, specific genes, exons or sequence stretches. Probes are 50-120 bp length of DNA or RNA with complementary sequences that bind to target regions to form hybridized DNA. This hybridized DNA is captured with the streptavidin-tagged magnetic beads , purified by magnetic pull-down, the target DNA is then eluted and used for library preparation and sequencing.
Here are some hints and tips to keep in mind when selecting your beads and designing your experiment:
- Hybrid capture-based target enrichment allows comprehensive analysis of variant types (single nucleotide polymorphisms, insertion/deletions, copy number variations, structural variations) by capturing large target regions in a single experiment. This is advantageous along with providing better resolution over conventional methods like PCR and molecular inversion probes (MIPs).
- Uniformly sheared genomic DNA positively influences downstream success of sequencing. Sonication is a preferred way of attaining homogeneity.
- Smaller fragments bind with greater specificity to probes over large fragments. Therefore, uniform shearing and fragment size selection is important for enhanced efficacy of an experiment.
- Uniformly synthesized probes provide better sequencing efficiency.
- Double-stranded probes maximize capture efficiency by providing more chances to capture a fragment.
- Pre-hybridization DNA amplification is recommended for samples of low integrity, such as clinical samples and aDNA.
- Hybridize the DNA and probe in a desalted environment to maximize specificity and efficiency.
- Probe-DNA hybridization is a critical step during which temperature and time are important factors. dsDNA denature to ssDNA at high temperature and convert to dsDNA at low temperature during which the DNA pairs with the biotinylated probes (refer to the manufacturer’s recommendations for best results).
- Gently vortex the probe-DNA mix every few hours for homogeneity and efficient hybridization. Certain genomic regions can be difficult to capture because of unfriendly or repeat sequences.
- Always wash away unbound DNA before eluting the targets
Unhybridized DNA can form a major contaminant in sequencing and can sometimes bind to streptavidin beads and increase false positives. Working with chilled buffers, increasing wash stringency, time and performing washes at room temperature can help to eliminate unhybridized DNA bound to streptavidin beads.
Reference.(1) He, H. et al. Integrated DNA and RNA extraction using magnetic beads from viral pathogens causing acute respiratory infections. Scientific reports, 7, 45199 (2017).