What is epitope binning?
Monoclonal antibodies (mAbs) specific to the same target protein are tested pairwise against all mAbs in a set to assess whether they block one another’s binding to a specific site of the antigen. The mAbs that block binding to the same epitope are “binned” together. This article discusses how SPR is used to characterize closely related bins of mAbs targeting the same epitope of the target protein and how the selection process is used to develop new vaccines, therapeutic antibodies, and diagnostic tools.
Understanding the need for epitope binning.
Epitope binning is used to characterize the binding of mAbs to a target protein, the antigen. Therapeutic mAbs are a large and growing part of the biopharmaceutical market, accounting for over 50% of therapeutic protein sales. It’s critical that mAbs with the appropriate affinity, specificity, and biophysical properties are selected during development of vaccines, therapeutic antibodies, and diagnostic tools.
Where does epitope binning fit in the antibody development process?
Early stage antibody drug development efforts generate many leads, and the best candidates must be chosen for further investigation. mAbs within the same bin often function similarly, so epitope bins can narrow down the choices to fewer candidates for investigators to choose from.
Although the antibodies within a bin bind to the same antigen, they may have different mechanisms of action, which is critical for treating some types of cancers and infectious diseases. Epitope diversity is also important to broaden intellectual property (IP) protection (1).
What are the analytical strategies for better informed antibody selection?
Antibodies have evolved from simpler mAbs to more complex biological entities including BITES, multispecifics, and conjugates. This increases the analytical burden and drives a need for more advanced analytical techniques. Since the early 1990s, biosensors based on surface plasmon resonance (SPR) have been used for epitope binning.
Epitope binning using conventional ELISAs, and radioimmunoassays (RIAs) require significant amounts of time to purify antibodies and develop labelling methods. Epitope binning assays using label-free biosensors allow a candidate molecule to be tested at relatively low cost and in an automated manner for efficient drug candidate optimization and identification of antibody pairs.
How is epitope binning performed?
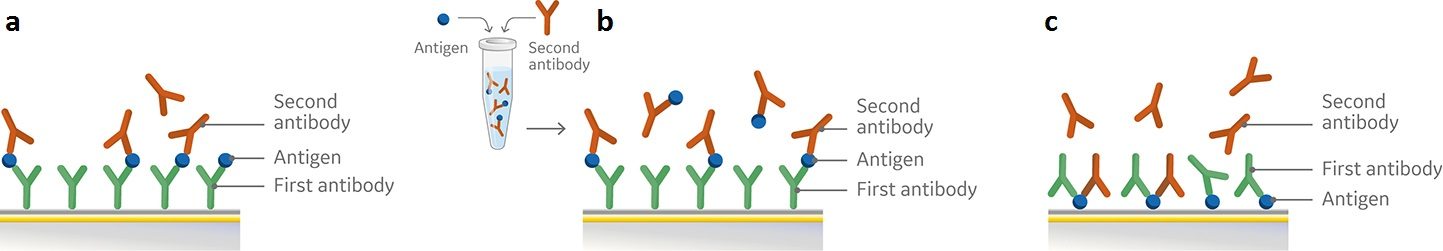
There are a range of formats for easy setup and running of an epitope binning assay with SPR. Common to all assay formats, including ELISA based approaches, are the use of an antigen and two antibodies that bind to the antigen. Biacore 1K, Biacore 8K and Biacore 8K+ systems combined with Biacore Insight Evaluation software and the new Biacore Insight Epitope Binning extension support three generic assay formats: Sandwich, premix, and tandem for epitope binning as predefined method protocols (Fig 1).
Fig 1. Schematic overview of set-up for pairwise epitope binning using a) sandwich assay, b) premix assay, and c) tandem assay. All three assay formats involve attachment of the first antibody (a, b) or the antigen (c) to the sensor chip surface followed by subsequent injections of either the antigen and second antibody in sequence (a), premixed (b), or the two antibodies (c).
Surface plasmon resonance (SPR) assay setup.
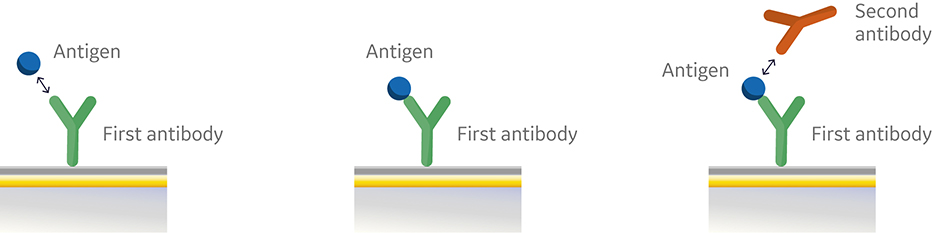
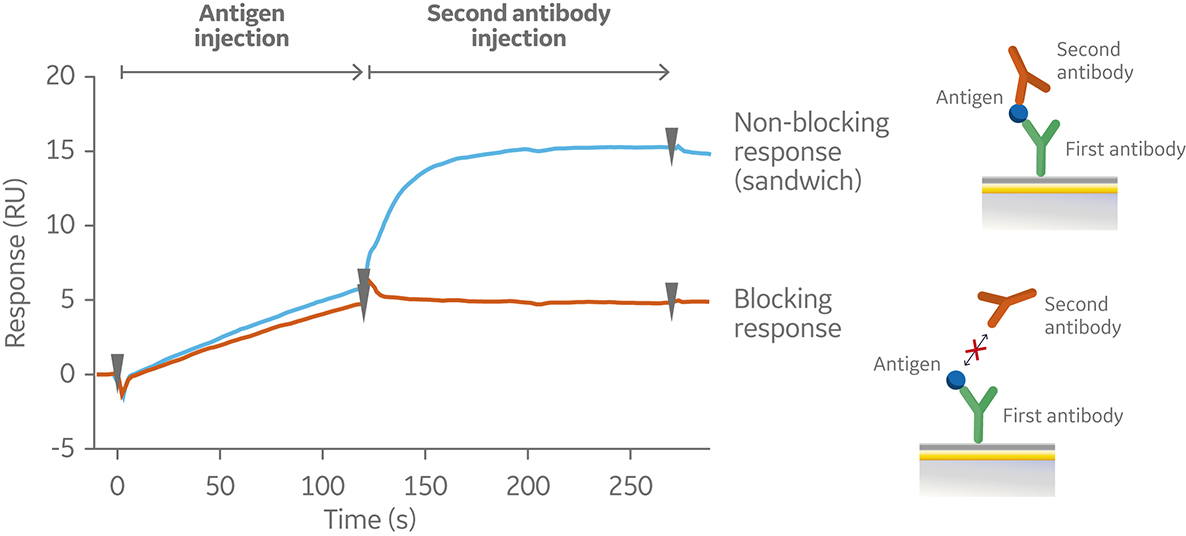
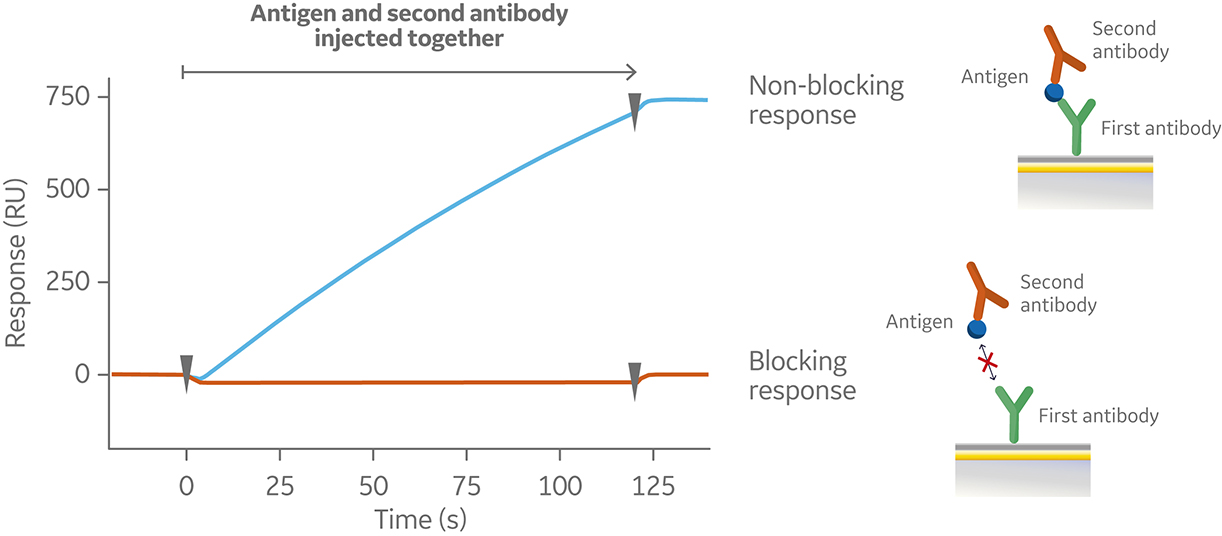
In the sandwich assay shown in Figure 2, the first antibody is captured or covalently attached to the surface of the sensor. The antigen is then injected and binds to the first antibody. This is followed by injection of the second antibody. The sensorgram in Figure 3 from 0 to 120 seconds shows the antigen binding to the immobilized first antibody. At 120 seconds the second antibody is injected. The second antibody either binds to the first antigen resulting in a “sandwich” binding response (blue) or is blocked from binding to the antigen by the first antibody (orange). If the second antibody can bind to the antigen in the presence of the first antibody, they have different epitopes and belong to different bins. If the second antibody is not able to bind to the antigen, both antibodies belong to the same bin.
Fig 2. Injection order of a sandwich assay.
Fig 3. Sensorgram illustrating the outcome of a sandwich assay with a non-blocking (blue) and blocking response (orange) from pairwise testing of two antibodies to an antigen. The first antibody was pre-immobilized to the sensor chip surface (not shown).
Fig 4. Injection order of a premix assay setup.
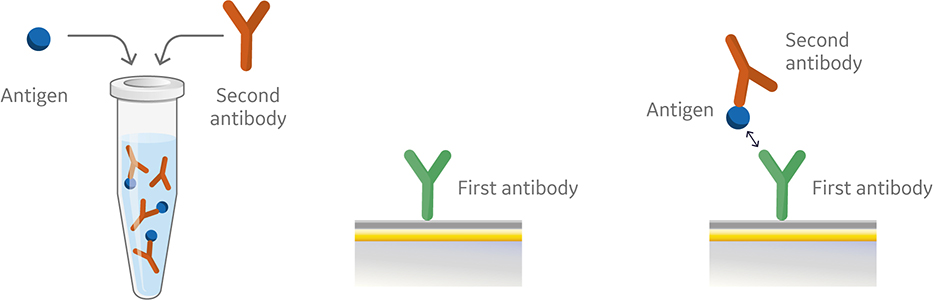
Like the sandwich assay, the first antibody in the premix assay is captured or covalently coupled to the surface of the sensor chip. However, in the premix assay the second antibody and antigen are mixed outside of the instrument using saturating conditions prior to injection (Fig 4). The antibody and the antigen are then injected into the instrument. Figure 5 shows a sensorgram from a premix assay.
Fig 5. Sensorgram from a premix assay illustrating a non-blocking (blue) and blocking interaction (orange) as a result of injection of premixed antigen and second antibody over a pre-immobilized first anitbody (not shown in sensorgram).
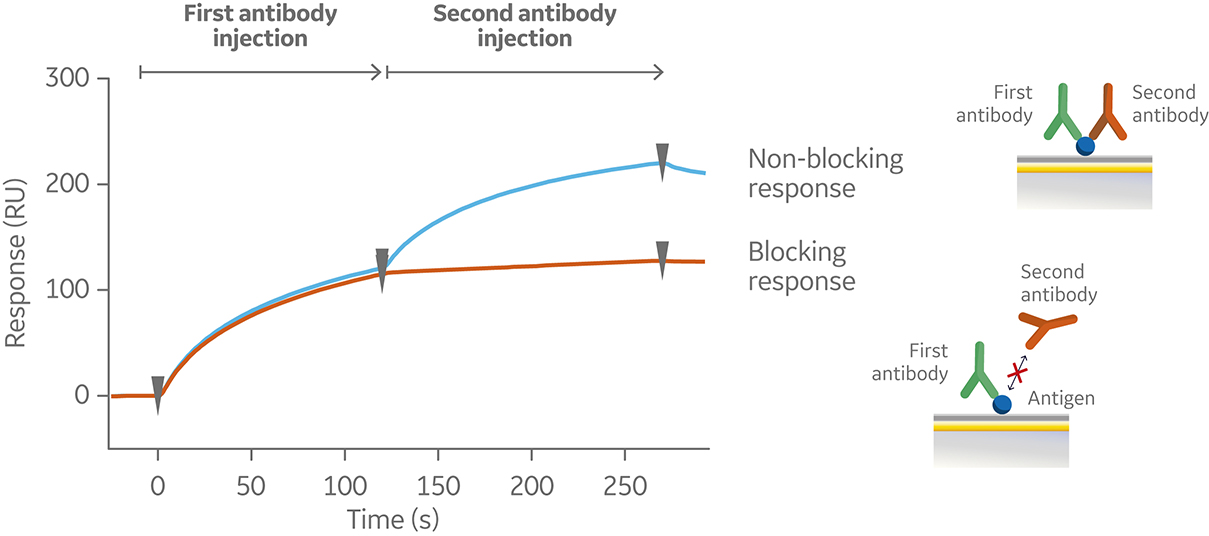
In a tandem assay, the antigen is captured or covalently coupled to the sensor surface. The first antibody is then injected at saturating antigen binding conditions followed by injection of the second antibody. A typical sensorgram for a tandem assay is shown in Figure 6.
Fig 6. Sensorgram from a tandem assay, showing a non-blocking (blue) and blocking (orange) response following subsequent injections of two antibodies against a pre-immobilized antigen (not shown in the sensorgram).
The different assay formats are complementary but have different advantages and disadvantages. A sandwich assay has a straightforward set up when regeneration conditions are known. The premix assay format is compatible with multivalent antigens and is often used if results from a sandwich assay need to be confirmed. However, the concentration of premix antibody must be in large excess over antigen concentration.
The tandem assay format is conceptually simple and is compatible with complex, multivalent antigen targets. It requires low amounts of antigen when immobilized and the first antibody must saturate all specific binding sites.
Given the latest advances of Biacore systems, the choice of assay design is defined by the molecules and reagents under investigation, not by technology ability.
Epitope binning vs. epitope mapping
Epitope binning tests the competitive relationships between different antibodies. Epitope mapping identifies exact binding sites on the antigen. This technique aims to locate specific amino acids or regions that allow the epitope to be recognized by the antibody.
Automating your epitope binning to find sweet spot bins.
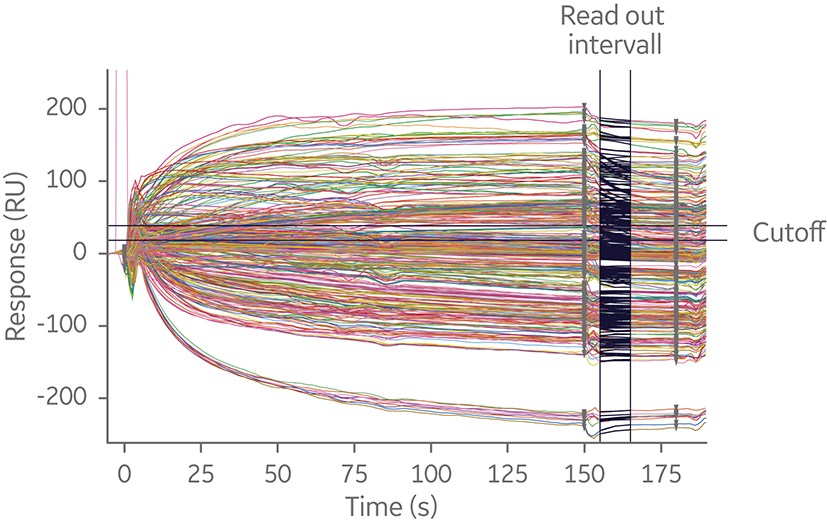
Within an assay format, runs may be performed in a symmetrical or asymmetrical fashion. For symmetrical assays, every pairwise variation of antibodies is tested against each other by switching the order in which the antibodies bind to their antigen. This approach may give a clear outcome, but for larger matrixes symmetrical runs result in a considerable amount of data in a short amount of time. This is because the number of interactions required scales geometrically with the number of antibodies in the test panel. For example, a 20 × 20 epitope binning analysis will generate 400 interactions (Fig 7).
Alternatively, asymmetric assays can be performed against a chosen number of antibodies representative of different epitope bins, or for example, already commercialized drugs whose bins should be avoided if new drugs are being developed. For example, an 8 × 200 assay would efficiently reduce the number of interactions to be followed up in a symmetrical run. A way to increase confidence in data for both symmetrical and asymmetrical runs is to compare results from an alternative assay format (for instance sandwich assay followed by a premix assay).
Fig 7. Sensorgram overlay of sandwich 20 × 20 SPR assay. The experiment utilized a capture antibody assay format to minimize time spent on assay development. Independent of where in the screening process epitope binning is performed, analyzing many interactions is time-consuming.
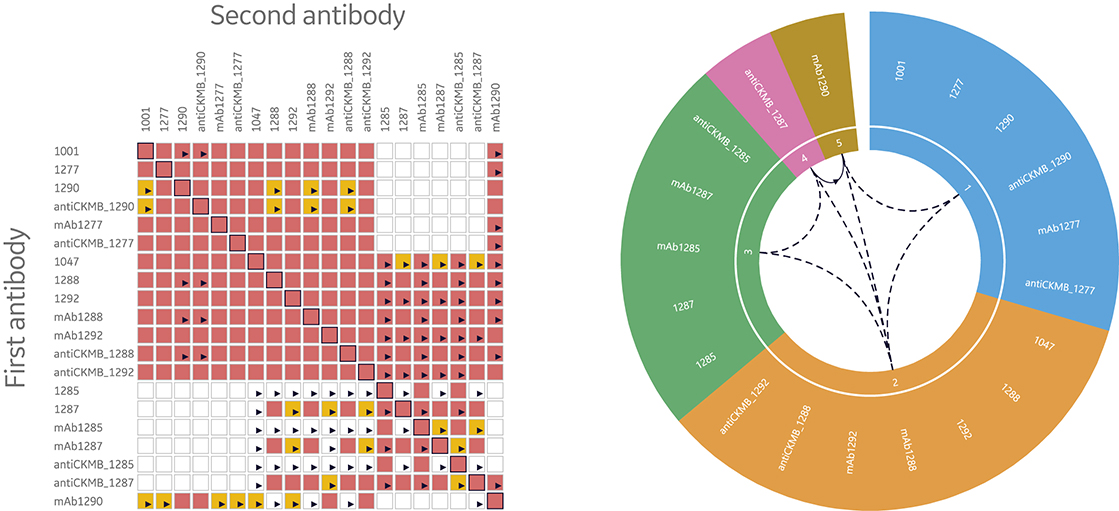
Consequently, it’s necessary to have tools to quickly interpret data by illustrating and evaluating epitope bins. Two ways to visualize data from epitope binning assays are heat maps and bin wheels, shown in Figure 8. The heat map used in Biacore Insight Epitope Binning extension presents an overview of the blocking (red), non- blocking (white), and uncertain antibody pairs (yellow). Thus, the heat map serves as a guide of uncertain results that need closer inspection. The bin wheel visualizes all detected bins in the current study by assigning antibodies with the same binning pattern the same color. Furthermore, antibodies with shared but not completely overlapping binning patterns are colored differently but aligned next to each other in the bin. Uni- and bi-directional behaviors between different bins are marked with arrows. Taken together, the bin wheel provides a comprehensive graphical representation of the results.
Fig 8. Heat maps and bin wheels are central features in the streamlined evaluation support provided in Biacore Insight Epitope Binning extension for more efficient data evaluation through the automatic analysis and visualization of bins for Biacore 8K series and Biacore T200 systems.
Overcome unstable binding of antigen to primary antibodies.
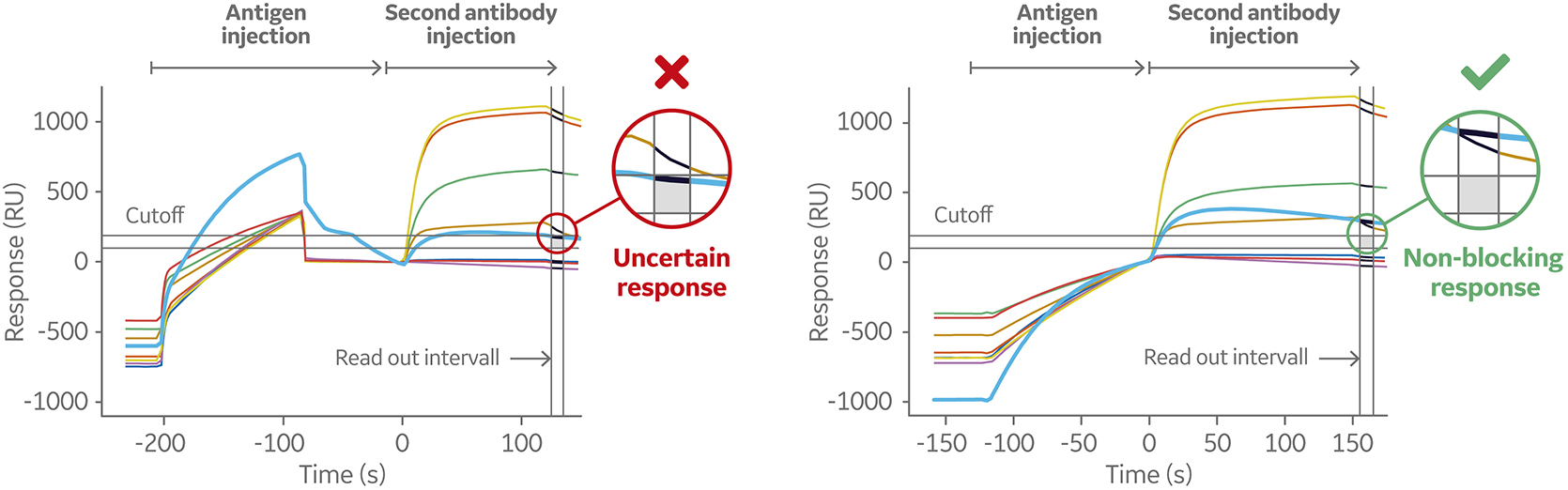
A common issue for sandwich and tandem assays in epitope binning using label free or ELISA techniques is low affinity of binding between the antigen and the first anitbody. This results in dissociation of the antigen during wash steps and underestimation of binding level of the second antibody, which leads to assignment of an antibody to the same bin when it’s not blocked at all.
To overcome this problem while using Biacore systems, the antigen and second antibody may be injected using the Dual command. It injects the two solutions in sequence with no intermediate washing steps. Thus, the dissociation of antigen before secondary antibody injection is minimized. This is one of the advantages of Biacore SPR systems over conventional technologies, such as ELISAs, for binning studies.
The Dual command allows for interpretation of binning data even for a fast dissociating antigen as it minimizes dissociation before the second antibody is injected.
Figure 9 shows the sensorgrams from an 8 × 1 sandwich assay using Biacore 8K system. Eight antibodies were pre-immobilized in channels 1 through 8, followed by one injection of antigen and one injection of second antibody. The assay was performed with separate injections (left) and repeated using the Dual injection command (right). One of the eight pre-immobilized antibodies displayed low affinity to the antigen.
Fig 9. Sensorgram (left) shows a weak-binding antigen to the first antibody, which may lead to underestimation of second antibody binding level. The sensorgram on the right shows the same sandwich assays set up using the Dual command. This injects the two solutions in sequence with no intermediate washing steps minimizing the dissociation of the antigen before the secondary antibody is injected.
Conclusion
A critical part of drug discovery and development efforts is finding and focusing on the most promising candidates. Epitope binning analysis using SPR systems gives investigators functionality information on therapeutic antibodies earlier in the development process. Biacore 8K and Biacore 8K+ systems combined with Biacore Insight Epitope Binning extension provides built in support and automated analysis tools for rapid evaluation of data from the most used epitope binning assay formats. This allows investigators to rapidly prioritize samples for further characterization studies and reach conclusions with confidence.
RELATED LITERATURE
Biacore reference list; a selection of publications presenting where and how SPR technology and Biacore systems have been applied.
FAQs
-
Why use surface plasmon resonance (SPR) for epitope binning?
SPR epitope binning is automated and can be performed without developing labelling methods. Biacore SPR systems provide tools to quickly interpret data by illustrating and evaluating epitope bins. The heat map used in Biacore Insight Epitope Binning extension serves as a guide of uncertain results that need closer inspection. The bin wheel visualizes all detected bins in the current study by assigning antibodies with the same binning pattern the same color. -
Can antibodies in the same bin function differently?
Yes. Although antibodies within a bin bind to the same antigen, they may have different mechanisms of action, which is critical for treating some types of cancers and infectious diseases. What are the benefits of SPR over traditional assays like ELISA?
Epitope binning using conventional ELISAs, and radioimmunoassays (RIAs) require significant amounts of time to purify antibodies and develop labelling methods. Epitope binning assays using label-free biosensors, including Biacore SPR systems, allow a candidate molecule to be tested at relatively low cost and in an automated manner for efficient drug candidate optimization and identification of antibody pairs.- What are “bins” in epitope binning?
Monoclonal antibodies (mAbs) specific to the same target protein are tested pairwise against all mAbs in a set to assess whether they block one another’s binding to a specific site of the antigen. The mAbs that block binding to the same epitope are “binned” together.